J Breast Cancer.
2012 Sep;15(3):265-272. 10.4048/jbc.2012.15.3.265.
Personalized Medicine in Breast Cancer: A Systematic Review
- Affiliations
-
- 1Department of Statistics and Actuarial Science, Soongsil University, Seoul, Korea.
- 2Department of Surgery, Yonsei University College of Medicine, Seoul, Korea. skim@yuhs.ac
- KMID: 2286451
- DOI: http://doi.org/10.4048/jbc.2012.15.3.265
Abstract
- The recent advent of "-omics" technologies have heralded a new era of personalized medicine. Personalized medicine is referred to as the ability to segment heterogeneous subsets of patients whose response to a therapeutic intervention within each subset is homogeneous. This new paradigm in healthcare is beginning to affect both research and clinical practice. The key to success in personalized medicine is to uncover molecular biomarkers that drive individual variability in clinical outcomes or drug responses. In this review, we begin with an overview of personalized medicine in breast cancer and illustrate the most encountered statistical approaches in the recent literature tailored for uncovering gene signatures.
Keyword
Figure
Reference
-
1. Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001. 7:201–204.
Article2. Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998. 279:1200–1205.
Article3. Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997. 277:301–306.
Article4. President's Council of Advisors on Science and Technolgy. Priorities for Personalized Medicine. 2008. Washington, DC: President's Council of Advisors on Science and Technolgy.5. Pfizer. Think Science Now Perspective. Approaches to Cancer Care: the Promise of Personalized Medicine. 2010. New York: Pfizer.6. U.S. National Institutes of Health, U.S. National Library of Medicine. Genetics home reference: Glossary. Accessed July 1st, 2012. http://ghr.nlm.nih.gov/glossary=personalizedmedicine.7. Meadows M. Genomics and personalized medicine. FDA Consum. 2005. 39:12–17.
Article8. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001. 69:89–95.9. Micheel C, Ball J. Institute of Medicine (U.S.). Committee on Qualification of Biomarkers and Surrogate Endpoints in Chronic Disease. Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease. 2010. Washington, DC: National Academies Press.10. NIH consensus conference. Treatment of early-stage breast cancer. JAMA. 1991. 265:391–395.11. Clark GM, Zborowski DM, Culbertson JL, Whitehead M, Savoie M, Seymour L, et al. Clinical utility of epidermal growth factor receptor expression for selecting patients with advanced non-small cell lung cancer for treatment with erlotinib. J Thorac Oncol. 2006. 1:837–846.
Article12. Saez RA, McGuire WL, Clark GM. Prognostic factors in breast cancer. Semin Surg Oncol. 1989. 5:102–110.
Article13. Nemoto T, Natarajan N, Bedwani R, Vana J, Murphy GP. Breast cancer in the medial half. Results of 1978 National Survey of the American College of Surgeons. Cancer. 1983. 51:1333–1338.
Article14. Fisher B, Bauer M, Wickerham DL, Redmond CK, Fisher ER, Cruz AB, et al. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer. An NSABP update. Cancer. 1983. 52:1551–1557.
Article15. Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989. 63:181–187.
Article16. Rosen PP, Groshen S, Kinne DW, Norton L. Factors influencing prognosis in node-negative breast carcinoma: analysis of 767 T1N0M0/T2N0M0 patients with long-term follow-up. J Clin Oncol. 1993. 11:2090–2100.
Article17. Carstens PH, Greenberg RA, Francis D, Lyon H. Tubular carcinoma of the breast. A long term follow-up. Histopathology. 1985. 9:271–280.
Article18. Clayton F. Pure mucinous carcinomas of breast: morphologic features and prognostic correlates. Hum Pathol. 1986. 17:34–38.
Article19. Ridolfi RL, Rosen PP, Port A, Kinne D, Miké V. Medullary carcinoma of the breast: a clinicopathologic study with 10 year follow-up. Cancer. 1977. 40:1365–1385.
Article20. Le Doussal V, Tubiana-Hulin M, Friedman S, Hacene K, Spyratos F, Brunet M. Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer. 1989. 64:1914–1921.
Article21. Rosen PP, Groshen S, Saigo PE, Kinne DW, Hellman S. Pathological prognostic factors in stage I (T1N0M0) and stage II (T1N1M0) breast carcinoma: a study of 644 patients with median follow-up of 18 years. J Clin Oncol. 1989. 7:1239–1251.
Article22. Neville AM, Bettelheim R, Gelber RD, Säve-Söderbergh J, Davis BW, Reed R, et al. The International (Ludwig) Breast Cancer Study Group. Factors predicting treatment responsiveness and prognosis in node-negative breast cancer. J Clin Oncol. 1992. 10:696–705.
Article23. Winstanley J, Cooke T, Murray GD, Platt-Higgins A, George WD, Holt S, et al. The long term prognostic significance of c-erbB-2 in primary breast cancer. Br J Cancer. 1991. 63:447–450.
Article24. Borg A, Tandon AK, Sigurdsson H, Clark GM, Fernö M, Fuqua SA, et al. HER-2/neu amplification predicts poor survival in node-positive breast cancer. Cancer Res. 1990. 50:4332–4337.25. Paterson MC, Dietrich KD, Danyluk J, Paterson AH, Lees AW, Jamil N, et al. Correlation between c-erbB-2 amplification and risk of recurrent disease in node-negative breast cancer. Cancer Res. 1991. 51:556–567.26. Clark GM, McGuire WL. Follow-up study of HER-2/neu amplification in primary breast cancer. Cancer Res. 1991. 51:944–948.27. Nixon AJ, Neuberg D, Hayes DF, Gelman R, Connolly JL, Schnitt S, et al. Relationship of patient age to pathologic features of the tumor and prognosis for patients with stage I or II breast cancer. J Clin Oncol. 1994. 12:888–894.
Article28. Albain KS, Allred DC, Clark GM. Breast cancer outcome and predictors of outcome: are there age differentials? J Natl Cancer Inst Monogr. 1994. 35–42.29. Daly MB, Clark GM, McGuire WL. Breast cancer prognosis in a mixed Caucasian-Hispanic population. J Natl Cancer Inst. 1985. 74:753–757.30. Elledge RM, Clark GM, Chamness GC, Osborne CK. Tumor biologic factors and breast cancer prognosis among white, Hispanic, and black women in the United States. J Natl Cancer Inst. 1994. 86:705–712.
Article31. Pierce L, Fowble B, Solin LJ, Schultz DJ, Rosser C, Goodman RL. Conservative surgery and radiation therapy in black women with early stage breast cancer. Patterns of failure and analysis of outcome. Cancer. 1992. 69:2831–2841.
Article32. Oldenhuis CN, Oosting SF, Gietema JA, de Vries EG. Prognostic versus predictive value of biomarkers in oncology. Eur J Cancer. 2008. 44:946–953.
Article33. Bentzen SM, Buffa FM, Wilson GD. Multiple biomarker tissue microarrays: bioinformatics and practical approaches. Cancer Metastasis Rev. 2008. 27:481–494.
Article34. Clark GM. Prognostic factors versus predictive factors: examples from a clinical trial of erlotinib. Mol Oncol. 2008. 1:406–412.
Article35. Phan JH, Moffitt RA, Stokes TH, Liu J, Young AN, Nie S, et al. Convergence of biomarkers, bioinformatics and nanotechnology for individualized cancer treatment. Trends Biotechnol. 2009. 27:350–358.
Article36. National Cancer Institute. Drug information: drugs approved for different types of cancer. Accessed July 1st, 2012. http://www.cancer.gov/cancertopics/druginfo/drug-page-index.37. National Cancer Institute. Drug information: drugs approved for breast cancer. Accessed July 1st, 2012. http://www.cancer.gov/cancertopics/druginfo/breastcancer.38. Grann VR, Troxel AB, Zojwalla NJ, Jacobson JS, Hershman D, Neugut AI. Hormone receptor status and survival in a population-based cohort of patients with breast carcinoma. Cancer. 2005. 103:2241–2251.
Article39. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005. 365:1687–1717.40. Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005. 353:1673–1684.
Article41. Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007. 369:29–36.
Article42. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002. 20:719–726.
Article43. Jones DS, Hofmann L, Quinn S. 21st Century Medicine: A New Model for Medical Education and Practice. 2010. Gig Harbor: The Institute for Functional Medicine;23–87.44. Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997. 336:1401–1408.
Article45. National Cancer Institute. BRCA1 and BRCA2: cancer risk and genetic testing. Accessed July 1st, 2012. http://www.cancer.gov/cancertopics/factsheet/Risk/BRCA.46. U.S. Food and Drug Administration. Drugs@FDA: FDA approved drug products. Accessed July 1st, 2012. http://www.accessdata.fda.gov/scripts/cder/drugsatfda/.47. Hornberger J, Cosler LE, Lyman GH. Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care. 2005. 11:313–324.48. Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006. 24:3726–3734.
Article49. Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004. 164:35–42.50. MammaPrint Agendia. Accessed July 1st, 2012. http://www.agendia.com/pages/mammaprint/21.php.51. Personalized Medicine Coalition. The Case for Personalized Medicine. 2011. 3rd ed. Washington, DC: Personalized Medicine Coalition.52. Catchpoole DR, Kennedy P, Skillicorn DB, Simoff S. The curse of dimensionality: a blessing to personalized medicine. J Clin Oncol. 2010. 28:e723–e724.
Article53. Cheng F, Cho SH, Lee JK. Multi-gene expression-based statistical approaches to predicting patients' clinical outcomes and responses. Methods Mol Biol. 2010. 620:471–484.
Article54. Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007. 104:13086–13091.
Article55. Smith SC, Baras AS, Dancik G, Ru Y, Ding KF, Moskaluk CA, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011. 12:137–143.
Article56. Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001. 98:10787–10792.
Article57. Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006. 10:515–527.
Article58. Efron B, Tibshirani R, Storey JD, Tusher V. Empirical bayes analysis of a microarray experiment. J Am Stat Assoc. 2001. 96:1151–1160.
Article59. Dudoit R, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat Sin. 2002. 12:111–139.60. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004. 3:Article 3.
Article61. Hu J, Wright FA. Assessing differential gene expression with small sample sizes in oligonucleotide arrays using a mean-variance model. Biometrics. 2007. 63:41–49.
Article62. Lönnstedt I, Speed T. Replicated microarray data. Stat Sin. 2002. 12:31–46.63. Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol. 2001. 8:37–52.
Article64. Newton MA, Noueiry A, Sarkar D, Ahlquist P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics. 2004. 5:155–176.
Article65. Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, et al. Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol. 2001. 8:625–637.
Article66. Wang S, Ethier S. A generalized likelihood ratio test to identify differentially expressed genes from microarray data. Bioinformatics. 2004. 20:100–104.
Article67. Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003. 19:1945–1951.
Article68. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999. 286:531–537.
Article69. Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci U S A. 2000. 97:262–267.
Article70. Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000. 16:906–914.
Article71. Mukherjee S, Tamayo P, Slonim D, Verri A, Golub T, Mesirov JP, et al. Support Vector Machine Classication of Microarray Data. 1998. Cambridge: Massachusetts Institute of Technology.72. West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001. 98:11462–11467.
Article73. Nguyen DV, Rocke DM. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics. 2002. 18:39–50.
Article74. Nagji AS, Cho SH, Liu Y, Lee JK, Jones DR. Multigene expression-based predictors for sensitivity to Vorinostat and Velcade in non-small cell lung cancer. Mol Cancer Ther. 2010. 9:2834–2843.
Article75. Culhane AC, Perrière G, Considine EC, Cotter TG, Higgins DG. Between-group analysis of microarray data. Bioinformatics. 2002. 18:1600–1608.
Article76. Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997. 30:1145–1159.
Article77. Hand DJ. Construction and Assessment of Classification Rules. 1997. Chichester: Wiley.78. Soukup M, Lee JK. Developing optimal prediction models for cancer classification using gene expression data. J Bioinform Comput Biol. 2004. 1:681–694.
Article79. Soukup M, Cho H, Lee JK. Robust classification modeling on microarray data using misclassification penalized posterior. Bioinformatics. 2005. 21:Suppl 1. i423–i430.
Article80. Buyse M, Sargent DJ, Grothey A, Matheson A, de Gramont A. Biomarkers and surrogate end points: the challenge of statistical validation. Nat Rev Clin Oncol. 2010. 7:309–317.
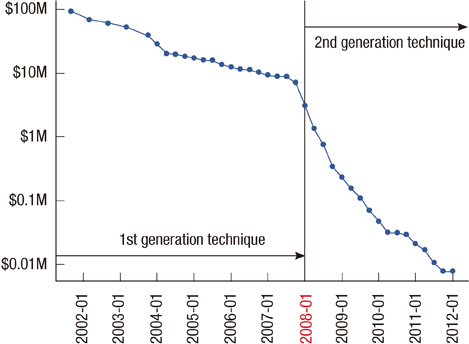
Article81. Wetterstrand KA. DNA sequencing costs: data from the NHGRI large scale genome sequencing program. Accessed July 1st, 2012. http://www.genome.gov/sequencingcosts/.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Breast Cancer Risk Prediction in Korean Women: Review and Perspectives on Personalized Breast Cancer Screening
- Diagnosis and Treatment of HER2-Positive Breast Cancer
- Overcoming the limitations of screening mammography in Japan and Korea: a paradigm shift to personalized breast cancer screening based on ultrasonography
- Psychometric Properties of Assessment Tools for Depression, Anxiety, Distress, and Psychological Problems in Breast Cancer Patients: A Systematic Review
- The Progress and Clinical Application of Breast Cancer Organoids