J Pathol Transl Med.
2022 Mar;56(2):73-80. 10.4132/jptm.2021.10.27.
Fatty acid synthetase expression in triple-negative breast cancer
- Affiliations
-
- 1Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
- 2Department of Surgery, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
- KMID: 2527191
- DOI: http://doi.org/10.4132/jptm.2021.10.27
Abstract
- Background
Triple-negative breast cancer (TNBC) has a relatively poor prognosis. Research has identified potential metabolic targets, including fatty acid metabolism, in TNBC. The absence of effective target therapies for TNBC led to exploration of the role of fatty acid synthetase (FASN) as a potential target for TNBC therapy. Here, we analyzed the expression of FASN, a representative lipid metabolism–related protein, and investigated the association between FASN expression and Ki-67 and the programmed death ligand 1 (PD-L1) biomarkers in TNBC.
Methods
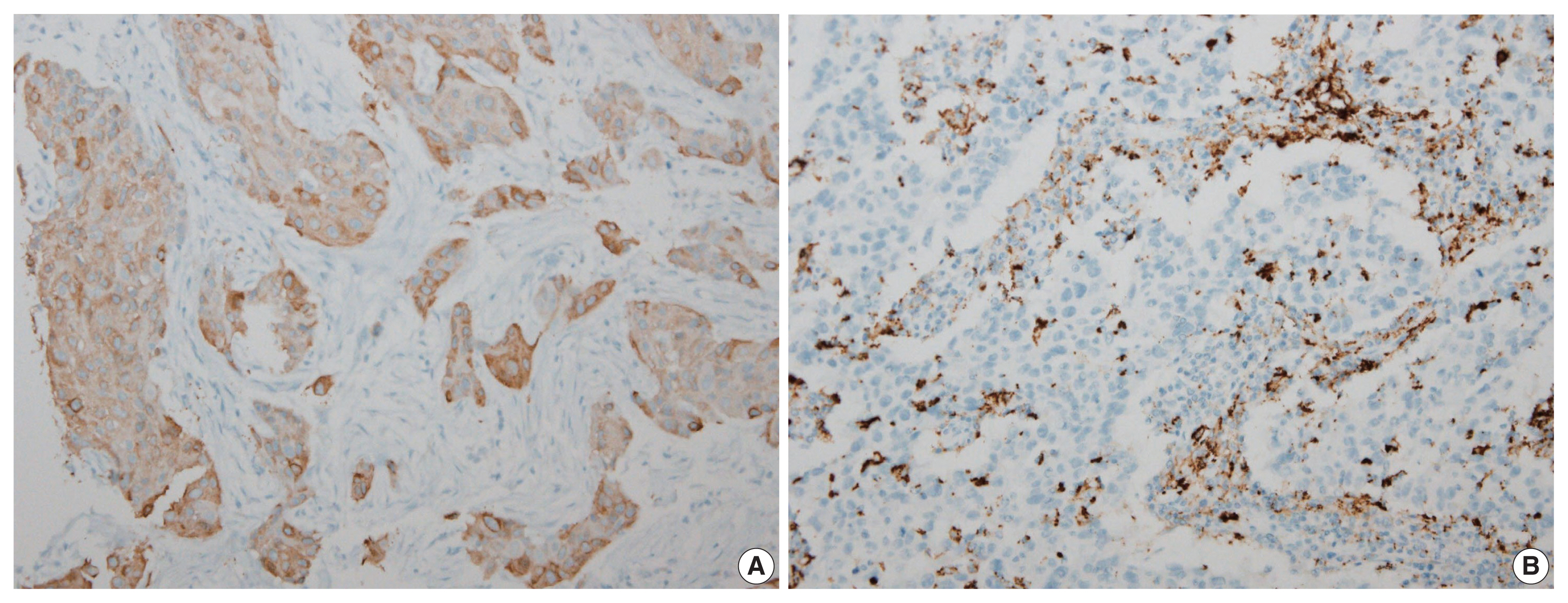
Immunohistochemical expression of FASN was analyzed in 166 patients with TNBC. For analytical purposes, patients with 0–1+ FASN staining were grouped as low-grade FASN and patients with 2–3+ FASN staining as high-grade FASN.
Results
FASN expression was observed in 47.1% of TNBC patients. Low and high expression of FASN was identified in 75.9% and 24.1%, respectively, and no statistically significant difference was found in T category, N category, American Joint Committee on Cancer stage, or recurrence rate between the low and high-FASN expression groups. Ki-67 proliferation level was significantly different between the low and high-FASN expression groups. FASN expression was significantly related to Ki-67 as the level increased. There was no significant difference in PD-L1 positivity between the low- and high-FASN expression groups.
Conclusions
We identified FASN expression in 166 TNBC patients. The Ki-67 proliferation index was positively correlated with FASN level, indicating higher proliferation activity as FASN increases. However, there was no statistical association with PD-L1 SP142, the currently FDA-approved assay, or FASN expression level.
Figure
Reference
-
References
1. Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer: current status and future directions. Ann Oncol. 2009; 20:1913–27.2. Diana A, Franzese E, Centonze S, et al. Triple-negative breast cancers: systematic review of the literature on molecular and clinical features with a focus on treatment with innovative drugs. Curr Oncol Rep. 2018; 20:76.
Article3. Lee KK, Kim JY, Jung JH, Park JY, Park HY. Clinicopathological feature and recurrence pattern of triple negative breast cancer. J Korean Surg Soc. 2010; 79:14–9.
Article4. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016; 13:674–90.
Article5. Marotti JD, de Abreu FB, Wells WA, Tsongalis GJ. Triple-negative breast cancer: next-generation sequencing for target identification. Am J Pathol. 2017; 187:2133–8.6. Lee SE, Park HY, Lim SD, Han HS, Yoo YB, Kim WS. Concordance of programmed death-ligand 1 expression between SP142 and 22C3/SP263 assays in triple-negative breast cancer. J Breast Cancer. 2020; 23:303–13.
Article7. Rugo HS, Loi S, Adams S, et al. PD-L1 immunohistochemistry assay comparison in atezolizumab plus nab-paclitaxel-treated advanced triple-negative breast cancer. J Natl Cancer Inst. 2021; 113:1733–43.8. Sun X, Wang M, Wang M, et al. Metabolic reprogramming in triple-negative breast cancer. Front Oncol. 2020; 10:428.
Article9. Gong Y, Ji P, Yang YS, et al. Metabolic-pathway-based subtyping of triple-negative breast cancer reveals potential therapeutic targets. Cell Metab. 2021; 33:51–64.
Article10. Cha YJ, Kim HM, Koo JS. Expression of lipid metabolism-related proteins differs between invasive lobular carcinoma and invasive ductal carcinoma. Int J Mol Sci. 2017; 18:232.
Article11. Sebastiani V, Visca P, Botti C, et al. Fatty acid synthase is a marker of increased risk of recurrence in endometrial carcinoma. Gynecol Oncol. 2004; 92:101–5.
Article12. Wang Y, Kuhajda FP, Li JN, et al. Fatty acid synthase (FAS) expression in human breast cancer cell culture supernatants and in breast cancer patients. Cancer Lett. 2001; 167:99–104.
Article13. Yang Y, Morin PJ, Han WF, et al. Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Exp Cell Res. 2003; 282:132–7.
Article14. Kusakabe T, Nashimoto A, Honma K, Suzuki T. Fatty acid synthase is highly expressed in carcinoma, adenoma and in regenerative epithelium and intestinal metaplasia of the stomach. Histopathology. 2002; 40:71–9.
Article15. Giro-Perafita A, Sarrats A, Perez-Bueno F, et al. Fatty acid synthase expression and its association with clinico-histopathological features in triple-negative breast cancer. Oncotarget. 2017; 8:74391–405.
Article16. Jiang W, Xing XL, Zhang C, et al. MET and FASN as prognostic biomarkers of triple negative breast cancer: a systematic evidence landscape of clinical study. Front Oncol. 2021; 11:604801.
Article17. Flores FH, Vinas G, Oliveras G, et al. Triple negative breast cancer: clinicopathologic characteristics and fatty acid synthase (FASN) expression as a potential target. Ann Oncol. 2014; 25(Suppl 4):IV60.
Article18. Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes: dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22:1736–47.19. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010; 134:e48–72.20. Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007; 131:18–43.21. Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008; 68:989–97.
Article22. Ma D, Jiang YZ, Xiao Y, et al. Integrated molecular profiling of young and elderly patients with triple-negative breast cancer indicates different biological bases and clinical management strategies. Cancer. 2020; 126:3209–18.
Article23. Li J, Holm J, Bergh J, et al. Breast cancer genetic risk profile is differentially associated with interval and screen-detected breast cancers. Ann Oncol. 2015; 26:517–22.
Article24. Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer. 2015; 15:248–54.
Article25. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–48.
Article26. Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015; 21:1688–98.
Article27. Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019; 9:176–98.
Article28. D’Ippolito E, Iorio MV. MicroRNAs and triple negative breast cancer. Int J Mol Sci. 2013; 14:22202–20.
Article29. Le Du F, Eckhardt BL, Lim B, et al. Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype? Oncotarget. 2015; 6:12890–908.
Article30. Hirai H, Tada Y, Nakaguro M, et al. The clinicopathological significance of the adipophilin and fatty acid synthase expression in salivary duct carcinoma. Virchows Arch. 2020; 477:291–9.
Article31. Kim S, Lee Y, Koo JS. Differential expression of lipid metabolism-related proteins in different breast cancer subtypes. PLoS One. 2015; 10:e0119473.
Article32. Giro-Perafita A, Rabionet M, Planas M, et al. EGCG-derivative G28 shows high efficacy inhibiting the mammosphere-forming capacity of sensitive and resistant TNBC models. Molecules. 2019; 24:1027.
Article33. Wang W, Bai L, Li W, Cui J. The lipid metabolic landscape of cancers and new therapeutic perspectives. Front Oncol. 2020; 10:605154.
Article34. Mrklic I, Capkun V, Pogorelic Z, Tomic S. Prognostic value of Ki-67 proliferating index in triple negative breast carcinomas. Pathol Res Pract. 2013; 209:296–301.
Article35. Ahmed ST, Ahmed AM, Musa DH, Sulayvani FK, Al-Khyatt M, Pity IS. Proliferative index (Ki67) for prediction in breast duct carcinomas. Asian Pac J Cancer Prev. 2018; 19:955–9.36. Miyashita M, Ishida T, Ishida K, et al. Histopathological subclassification of triple negative breast cancer using prognostic scoring system: five variables as candidates. Virchows Arch. 2011; 458:65–72.
Article37. Nielsen TO, Leung SC, Rimm DL, et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst. 2021; 113:808–19.
Article38. Zong Y, Zhu L, Wu J, et al. Progesterone receptor status and Ki-67 index may predict early relapse in luminal B/HER2 negative breast cancer patients: a retrospective study. PLoS One. 2014; 9:e95629.
Article39. Wang W, Wu J, Zhang P, et al. Prognostic and predictive value of Ki-67 in triple-negative breast cancer. Oncotarget. 2016; 7:31079–87.
Article40. Pan Y, Yuan Y, Liu G, Wei Y. P53 and Ki-67 as prognostic markers in triple-negative breast cancer patients. PLoS One. 2017; 12:e0172324.
Article41. Mavratzas A, Seitz J, Smetanay K, Schneeweiss A, Jager D, Fremd C. Atezolizumab for use in PD-L1-positive unresectable, locally advanced or metastatic triple-negative breast cancer. Future Oncol. 2020; 16:4439–53.
Article42. Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002; 8:793–800.
Article43. Peg V, Lopez-Garcia MA, Comerma L, et al. PD-L1 testing based on the SP142 antibody in metastatic triple-negative breast cancer: summary of an expert round-table discussion. Future Oncol. 2021; 17:1209–18.44. Li Y, Vennapusa B, Chang CW, et al. Prevalence study of PD-L1 SP142 assay in metastatic triple-negative breast cancer. Appl Immunohistochem Mol Morphol. 2021; 29:258–64.
Article45. Al-Jussani GN, Dabbagh TZ, Al-Rimawi D, Sughayer MA. Expression of PD-L1 using SP142 CDx in triple negative breast cancer. Ann Diagn Pathol. 2021; 51:151703.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinicopathologic Characteristics and Prognosis of Early Stage Triple Negative Breast Cancer: Comparison with Non-triple Negative Group
- Molecular Classification of Triple-Negative Breast Cancer
- Fear of Cancer Recurrence and Unmet Needs in Triple Negative Breast Cancer Survivors
- Overexpression of Cell Cycle Progression Inhibitor Geminin is Associated with Tumor Stem-Like Phenotype of Triple-Negative Breast Cancer
- Serum Phospholipid Fatty Acids in Benign Breast Tumor and Breast Cancer