J Bacteriol Virol.
2019 Dec;49(4):162-175. 10.4167/jbv.2019.49.4.162.
Analysis of Intestinal Mucosal Microbiome Changes before and after Chemoradiation in Locally Advanced Rectal Cancer Patients
- Affiliations
-
- 1Dasa Geriatric Hospital, Daegu, Korea.
- 2Department of Surgery, School of Medicine, Keimyung University and Dongsan Medical Center, Daegu, Korea. sabiston0000@hanmail.net
- 3Department of Immunology, School of Medicine, Keimyung University, Daegu, Korea.
- KMID: 2468023
- DOI: http://doi.org/10.4167/jbv.2019.49.4.162
Abstract
- PURPOSE
Dysbiosis of gut microbiota has been reported to participate in the pathogenesis of colorectal cancer, but changes in microbiota due to radiotherapy have not been studied. In this study, we tried to elucidate the changes in the microbiome in rectal cancer after chemoradiotherapy using RNA sequencing analysis.
MATERIALS AND METHODS
We included 11 pairs of human rectal cancer tissues before and after irradiation between August 2016 and December 2017 and performed RNA sequencing analysis. Mapped reads to human reference genomes were used for pair-wise transcriptome comparisons, and unmapped (non-human) reads were then mapped to bacterial marker genes using PathSeq.
RESULTS
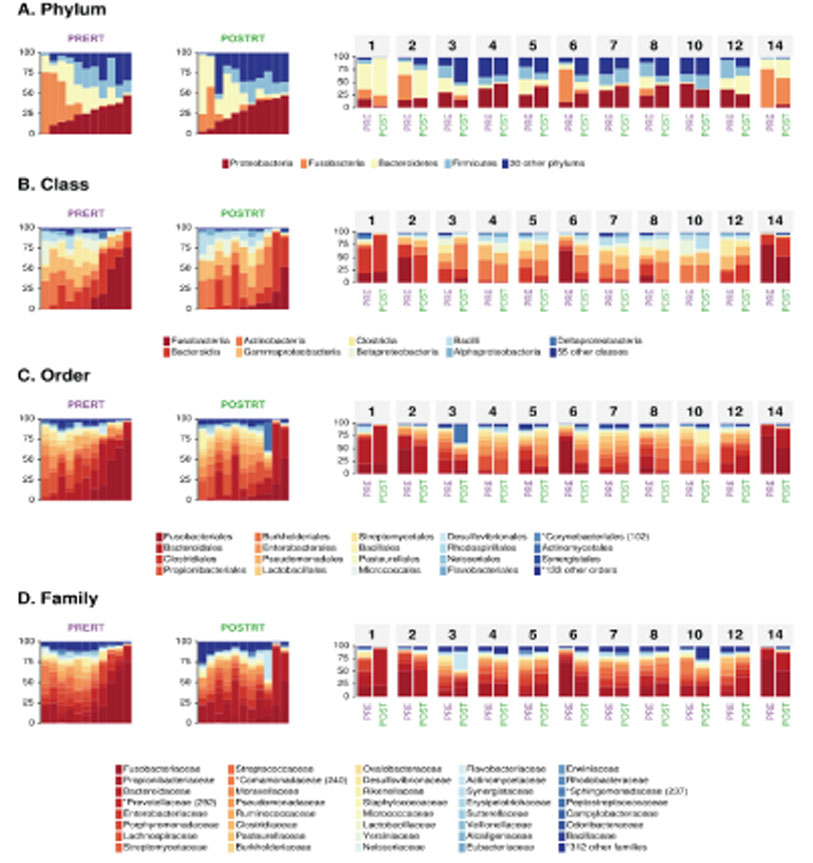
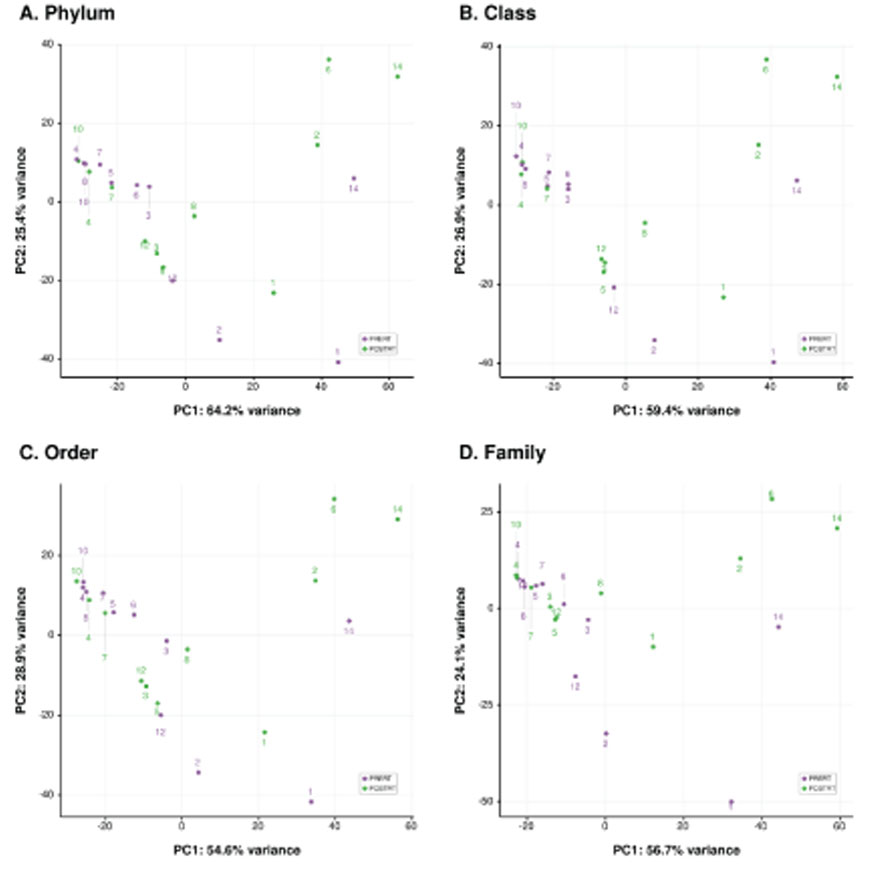
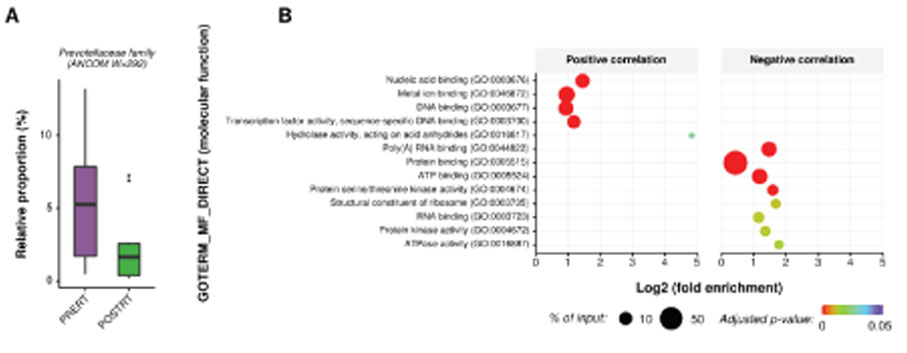
At microbiome level, interindividual variability of mucosal microbiota was greater than the change in microbial composition during radiotherapy. This indicates that rapid homeostatic recovery of the mucosal microbial composition takes place short after radiotherapy. At single microbe level, Prevotella and Fusobacterium, which were identified as important causative microbes of the initiation and progression of rectal cancer were decreased by radiotherapy. Moreover, changes in Prevotella were associated with changes in the human transcriptome of rectal cancer. We also found that there was a gene cluster that increased and decreased in association with changes in microbial composition by chemoradiation.
CONCLUSION
This study revealed changes in tumor-associated microbial community by irradiation in rectal cancer. These findings can be used to develop a new treatment strategy of neoadjuvant therapy for locally advanced rectal cancer by overcoming radio-resistance or facilitating radio-sensitivity.
Keyword
MeSH Terms
Figure
Reference
-
1. Swedish Rectal Cancer Trial. Improved survival with preoperative radiotherapy in resectable rectal cancer. N Engl J Med. 1997; 336:980–987.2. Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N Engl J Med. 2001; 345:638–646.
Article3. Bosset JF, Collette L, Calais G, Mineur L, Maingon P, Radosevic-Jelic L, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006; 355:1114–1123.
Article4. Han YD, Kim WR, Park SW, Cho MS, Hur H, Min BS, et al. Predictors of Pathologic Complete Response in Rectal Cancer Patients Undergoing Total Mesorectal Excision After Preoperative Chemoradiation. Medicine (Baltimore). 2015; 94:e1971.
Article5. Demaria S, Ng B, Devitt ML, Babb JS, Kawashima N, Liebes L, et al. Ionizing radiation inhibition of distant untreated tumors (abscopal effect) is immune mediated. Int J Radiat Oncol Biol Phys. 2004; 58:862–870.
Article6. Vatner RE, Cooper BT, Vanpouille-Box C, Demaria S, Formenti SC. Combinations of immunotherapy and radiation in cancer therapy. Front Oncol. 2014; 4:325.
Article7. Tuyaerts S, Van Nuffel AMT, Naert E, Van Dam PA, Vuylsteke P, De Caluwé A, et al. PRIMMO study protocol: a phase II study combining PD-1 blockade, radiation and immunomodulation to tackle cervical and uterine cancer. BMC Cancer. 2019; 19:506.
Article8. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017; 17:271–285.
Article9. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013; 342:967–970.
Article10. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015; 350:1084–1089.
Article11. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120.
Article12. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21.
Article13. Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RG, Getz G, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011; 29:393–396.
Article14. Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, Peddada SD. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015; 26:27663.
Article15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550.
Article16. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57.
Article17. Team RC. R: A language and environment for statistical computing. . Vienna, Austria: R Foundation for Statstical Computing;2018.18. Wickham H. Ggplot2: elegant graphics for data analysis. New York: Springer-Verlag;2016.19. Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, Moser AR, et al. Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997; 57:812–814.20. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010; 107:11537–11542.
Article21. Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006; 12:782–786.
Article22. Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015; 6:20.
Article23. Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013; 66:462–470.
Article24. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012; 10:575–582.
Article25. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012; 10:717–725.
Article26. Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial Biofilms in Colorectal Cancer Initiation and Progression. Trends Mol Med. 2017; 23:18–30.
Article27. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015; 350:1079–1084.
Article28. Kim YS, Kim J, Park SJ. High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe. 2015; 33:1–7.
Article29. Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, Gershovich K, Sabo E, Nevelsky A, et al. Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut. 2018; 67:97–107.
Article30. Wang A, Ling Z, Yang Z, Kiela PR, Wang T, Wang C, et al. Gut microbial dysbiosis may predict diarrhea and fatigue in patients undergoing pelvic cancer radiotherapy: a pilot study. PLoS One. 2015; 10:e0126312.
Article31. Nam YD, Kim HJ, Seo JG, Kang SW, Bae JW. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS One. 2013; 8:e82659.
Article32. Manichanh C, Varela E, Martinez C, Antolin M, Llopis M, Doré J, et al. The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. Am J Gastroenterol. 2008; 103:1754–1761.
Article33. Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017; 66:633–643.
Article34. Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteriain the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002; 68:3401–3407.
Article35. Ranawat P, Rawat S. Radiation resistance in thermophiles: mechanisms and applications. World J Microbiol Biotechnol. 2017; 33:112.
Article36. Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018; 3:1255–1265.
Article37. Jalanka J, Salonen A, Salojärvi J, Ritari J, Immonen O, Marciani L, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015; 64:1562–1568.
Article38. Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016; 11:e0152126.
Article39. Xu K, Jiang B. Analysis of Mucosa-Associated Microbiota in Colorectal Cancer. Med Sci Monit. 2017; 23:4422–4430.
Article40. Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, et al. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS One. 2016; 11:e0155362.
Article41. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013; 14:195–206.
Article42. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015; 1:653–661.
Article43. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011; 6:e16393.
Article44. Wu D, Wu P, Huang Q, Liu Y, Ye J, Huang J. Interleukin-17: a promoter in colorectal cancer progression. Clin Dev Immunol. 2013; 2013:436307.
Article45. Liu J, Duan Y, Cheng X, Chen X, Xie W, Long H, et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem Biophys Res Commun. 2011; 407:348–354.
Article46. Wang K. Targeting IL-17 for cancer-associated inflammation and immunity. J Immunol. 2017; 198:66.5.47. Crawford PA, Gordon JI. Microbial regulation of intestinal radiosensitivity. Proc Natl Acad Sci U S A. 2005; 102:13254–13259.
Article48. Ferreira MR, Muls A, Dearnaley DP, Andreyev HJ. Microbiota and radiation-induced bowel toxicity: lessons from inflammatory bowel disease for the radiation oncologist. Lancet Oncol. 2014; 15:e139–e147.
Article49. Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D, et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med. 2017; 9:448–461.
Article50. Nejdfors P, Ekelund M, Weström BR, Willén R, Jeppsson B. Intestinal permeability in humans is increased after radiation therapy. Dis Colon Rectum. 2000; 43:1582–1587. discussion 1587-8.
Article51. Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007; 117:2197–2204.
Article52. Goubet AG, Daillère R, Routy B, Derosa L, M Roberti P, Zitvogel L. The impact of the intestinal microbiota in therapeutic responses against cancer. C R Biol. 2018; 341:284–289.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Outcome of Preoperative Chemoradiation to Locally Advanced Rectal Cancer
- How to Achieve a Higher Pathologic Complete Response in Patients With Locally Advanced Rectal Cancer Who Receive Preoperative Chemoradiation Therapy
- The Effects and Surgical Morbidity of Preoperative Combined Chemoradiotherapy for Locally Advanced Rectal Cancer
- Disadvantages of Preoperative Chemoradiation in Rectal Cancer
- Imaging Diagnosis of Locally Advanced Rectal Cancer: Tumor Staging before and after Preoperative Chemoradiotherapy