Anat Cell Biol.
2019 Dec;52(4):469-477. 10.5115/acb.19.048.
Big data differential analysis of microglial cell responses in neurodegenerative diseases
- Affiliations
-
- 1Department of Anatomy and Cell Biology, Dong-A University College of Medicine, Busan, Korea.
- 2Department of Medicine, Graduate School, Dong-A University, Busan, Korea.
- 3Department of Anesthesiology and Pain Medicine, Kosin University College of Medicine, Busan, Korea. dmadjejs@naver.com
- KMID: 2466701
- DOI: http://doi.org/10.5115/acb.19.048
Abstract
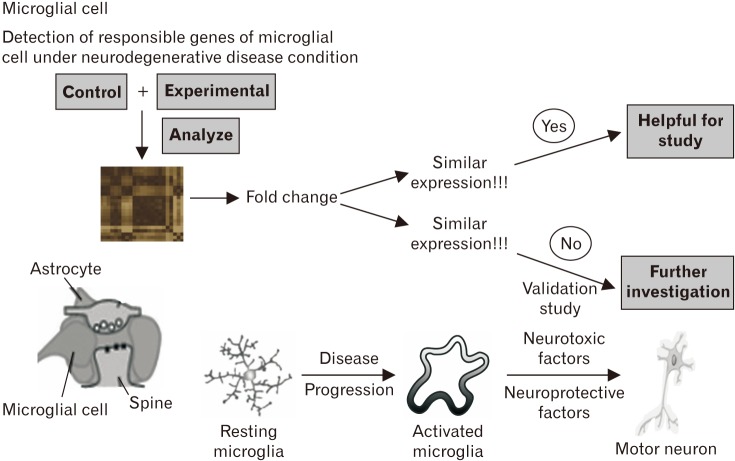
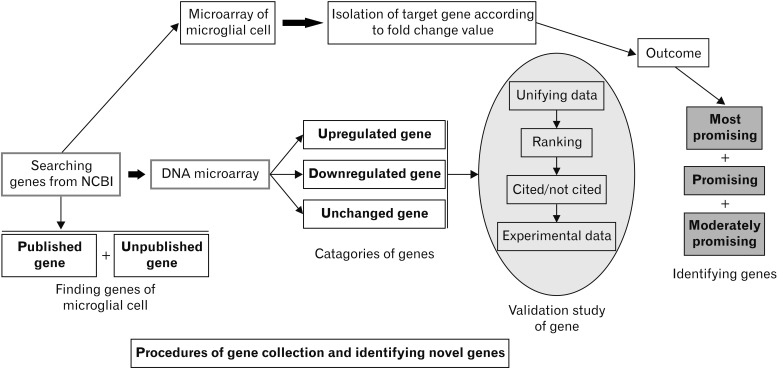
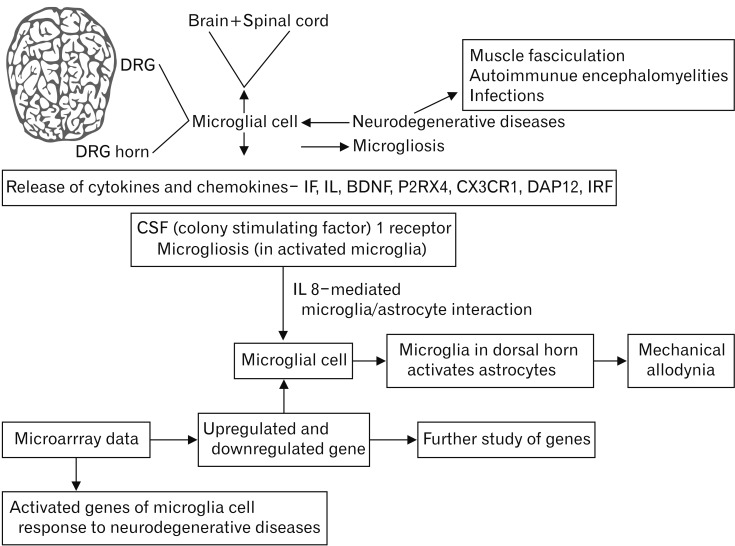
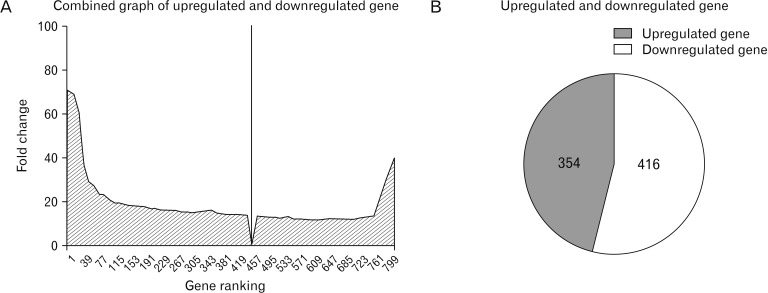
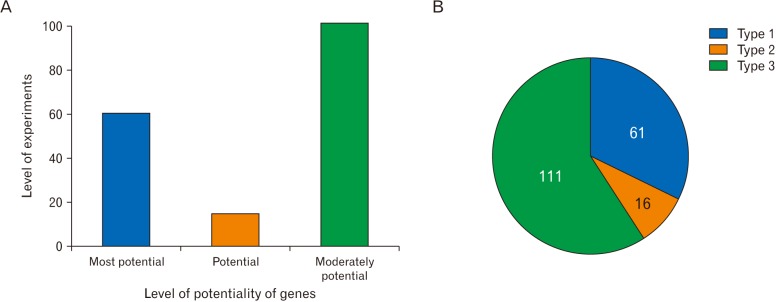
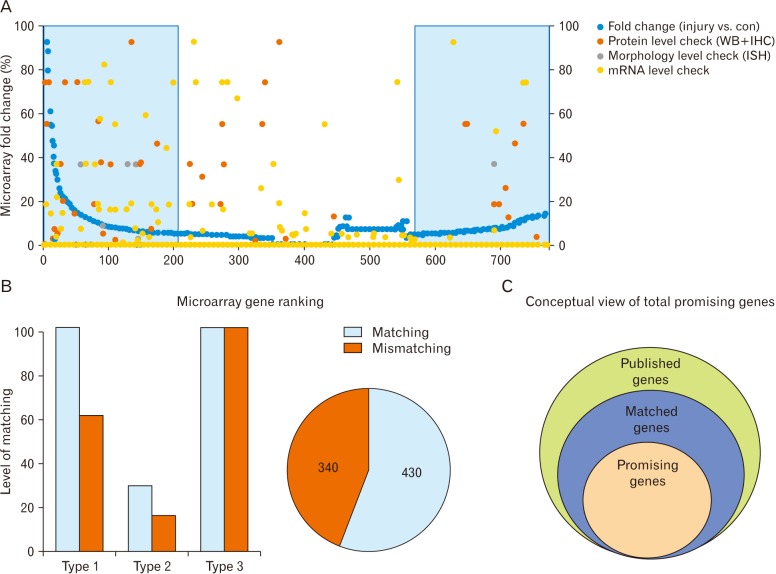
- Microarray technology has become an indispensable tool for monitoring the levels of gene expression in a given organism through organization, analysis, interpretation, and utilization of biological sequences. Importantly, preliminary microarray gene expression differs from experimentally validated gene expression. Generally, microarray analysis of gene expression in microglial cells is used to identify genes in the brain and spinal cord that are responsible for the onset of neurodegenerative diseases; these genes are either upregulated or downregulated. In the present study, 770 genes identified in prior publications, including experimental studies, were analyzed to determine whether these genes encode novel disease genes. Among the genes published, 340 genes were matched among multiple publications, whereas 430 genes were mismatched; the matched genes were presumed to have the greatest likelihood of contributing to neurodegenerative diseases and thus to be potentially useful target genes for treatment of neurodegenerative diseases. In protein and mRNA expression studies, matched and mismatched genes showed 99% and 97% potentiality, respectively. In addition, some genes identified in microarray analyses were significantly different from those in experimentally validated expression patterns. This study identified novel genes in microglial cells through comparative analysis of published microarray and experimental data on neurodegenerative diseases.
MeSH Terms
Figure
Cited by 1 articles
-
Camillo Golgi (1843 –1926): scientist extraordinaire and pioneer figure of modern neurology
Sanjib Kumar Ghosh
Anat Cell Biol. 2020;53(4):385-392. doi: 10.5115/acb.20.196.
Reference
-
1. Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000; 7:819–837. PMID: 11382364.2. Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol. 2001; 8:37–52. PMID: 11339905.3. Wake H, Moorhouse AJ, Miyamoto A, Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci. 2013; 36:209–217. PMID: 23260014.4. Trapp BD, Wujek JR, Criste GA, Jalabi W, Yin X, Kidd GJ, Stohlman S, Ransohoff R. Evidence for synaptic stripping by cortical microglia. Glia. 2007; 55:360–368. PMID: 17136771.5. Yamada J, Nakanishi H, Jinno S. Differential involvement of perineuronal astrocytes and microglia in synaptic stripping after hypoglossal axotomy. Neuroscience. 2011; 182:1–10. PMID: 21435379.6. Tsuda M, Inoue K, Salter MW. Neuropathic pain and spinal microglia: a big problem from molecules in “small” glia. Trends Neurosci. 2005; 28:101–107. PMID: 15667933.7. Inoue K, Tsuda M. Microglia in neuropathic pain: cellular and molecular mechanisms and therapeutic potential. Nat Rev Neurosci. 2018; 19:138–152. PMID: 29416128.8. Carson MJ. Microglia as liaisons between the immune and central nervous systems: functional implications for multiple sclerosis. Glia. 2002; 40:218–231. PMID: 12379909.9. Sweitzer SM, White KA, Dutta C, DeLeo JA. The differential role of spinal MHC class II and cellular adhesion molecules in peripheral inflammatory versus neuropathic pain in rodents. J Neuroimmunol. 2002; 125:82–93. PMID: 11960644.10. Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000; 25:294–297. PMID: 10888876.11. Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PLoS One. 2011; 6:e16266. PMID: 21283692.12. Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, Niwa J, Tanaka F, Doyu M, Yoshida M, Hashizume Y, Sobue G. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005; 57:236–251. PMID: 15668976.13. Thacker MA, Clark AK, Bishop T, Grist J, Yip PK, Moon LD, Thompson SW, Marchand F, McMahon SB. CCL2 is a key mediator of microglia activation in neuropathic pain states. Eur J Pain. 2009; 13:263–272. PMID: 18554968.14. Wada R, Tifft CJ, Proia RL. Microglial activation precedes acute neurodegeneration in Sandhoff disease and is suppressed by bone marrow transplantation. Proc Natl Acad Sci U S A. 2000; 97:10954–10959. PMID: 11005868.15. Lehesjoki AE, Koskiniemi M. Progressive myoclonus epilepsy of Unverricht-Lundborg type. Epilepsia. 1999; 40(Suppl 3):23–28.16. Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M, Pedotti R, Heller R, Oksenberg JR, Steinman L. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001; 294:1731–1735. PMID: 11721059.17. Lassmann H, Bradl M. Multiple sclerosis: experimental models and reality. Acta Neuropathol. 2017; 133:223–244. PMID: 27766432.18. Sinclair C, Mirakhur M, Kirk J, Farrell M, McQuaid S. Up-regulation of osteopontin and alphaBeta-crystallin in the normalappearing white matter of multiple sclerosis: an immunohistochemical study utilizing tissue microarrays. Neuropathol Appl Neurobiol. 2005; 31:292–303. PMID: 15885066.19. Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001; 6:293–301. PMID: 11326297.20. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003; 34:267–273. PMID: 12808457.21. Brown CW, Houston-Hawkins DE, Woodruff TK, Matzuk MM. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat Genet. 2000; 25:453–457. PMID: 10932194.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Activated microglial cells synthesize and secrete AGE-albumin
- Enhancement of Nitric Oxide Production by Corticotropin-releasing Hormone (CRH) in Murine Microglial Cells, BV2
- Antineuroinflammatory Effects of 7,3’,4’-Trihydroxyisoflavone in Lipopolysaccharide-Stimulated BV2 Microglial Cells through MAPK and NF-κB Signaling Suppression
- Gene Expression Analysis of Murine Primary Microglia Stimulated with LPS using Microarray
- Big Data: An Overview and Its Applications in Medicine and Aviation