Int J Stem Cells.
2019 Nov;12(3):463-473. 10.15283/ijsc19007.
Role of a 19S Proteasome Subunit- PSMD10(Gankyrin) in Neurogenesis of Human Neural Progenitor Cells
- Affiliations
-
- 1Advance Centre for Treatment, Research and Education in Cancer, Tata Memorial Centre, Kharghar, Navi Mumbai, India. vprasanna@actrec.gov.in
- 2Faculty of Biology, Technion – Israel Institute of Technology, Haifa, Israel.
- 3Homi Bhabha National Institute, BARC Training School Complex, Mumbai, Maharashtra, India.
- 4Department of Pathology, University of Massachusetts Medical School, Worcester, MA, USA.
- 5Department of Physics, Bose Institute, Kolkata, India. soumen@jcbose.ac.in
- KMID: 2465887
- DOI: http://doi.org/10.15283/ijsc19007
Abstract
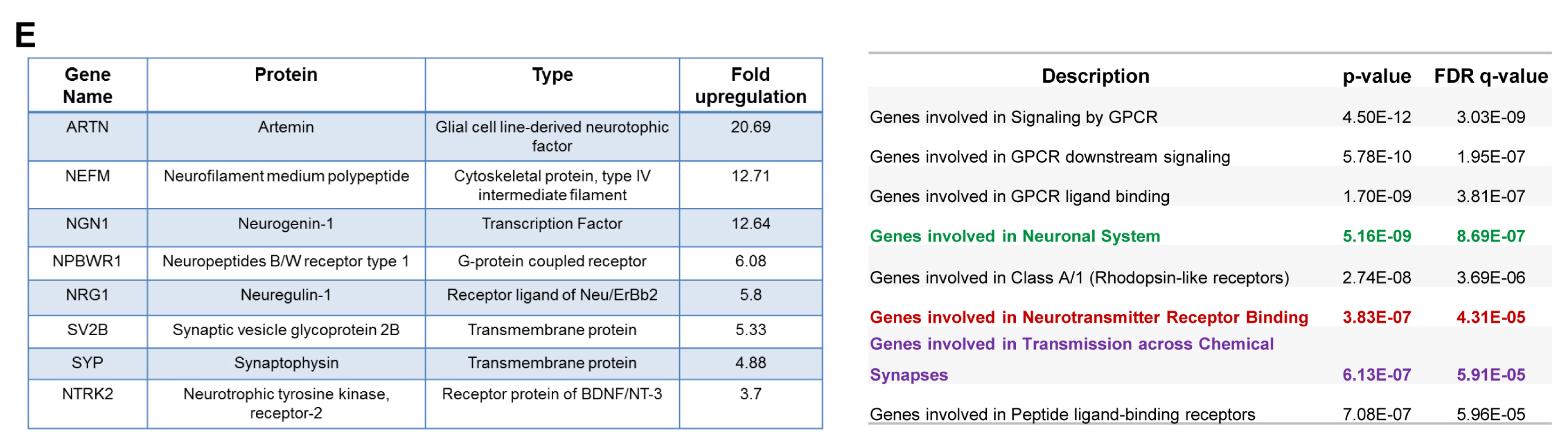
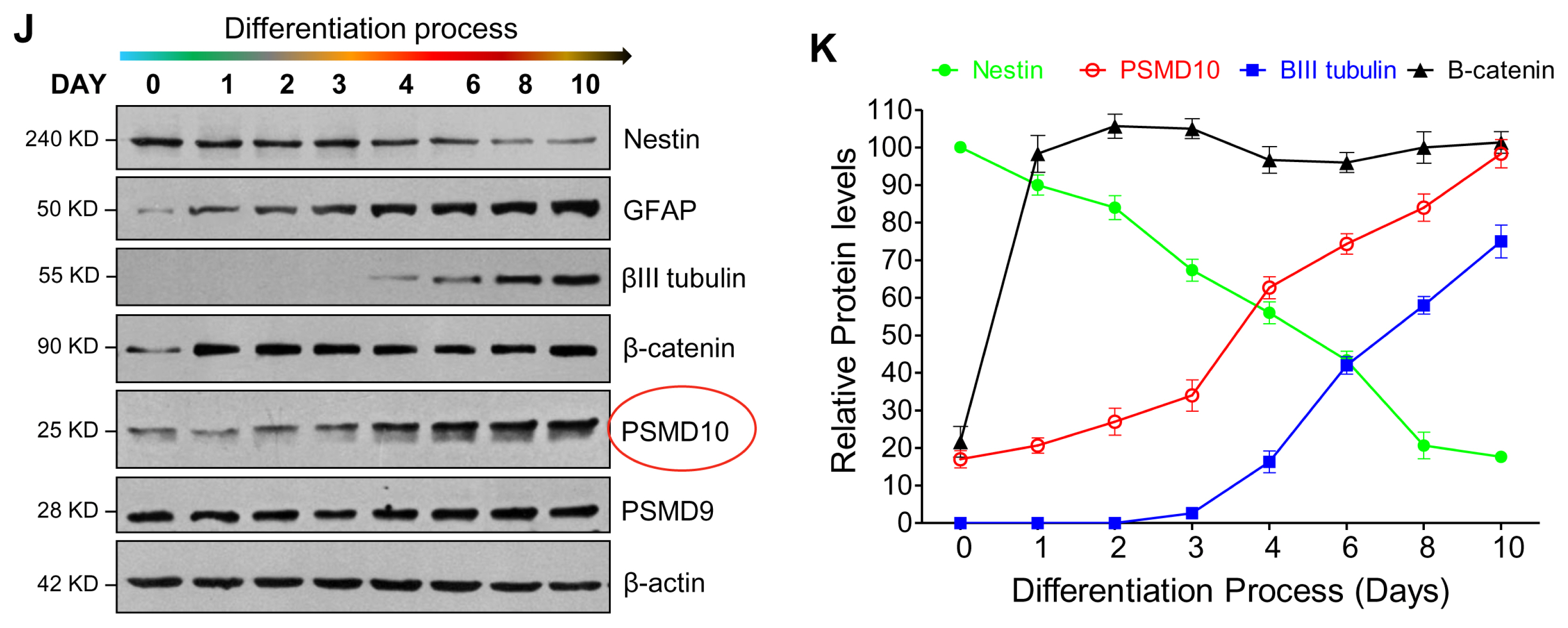
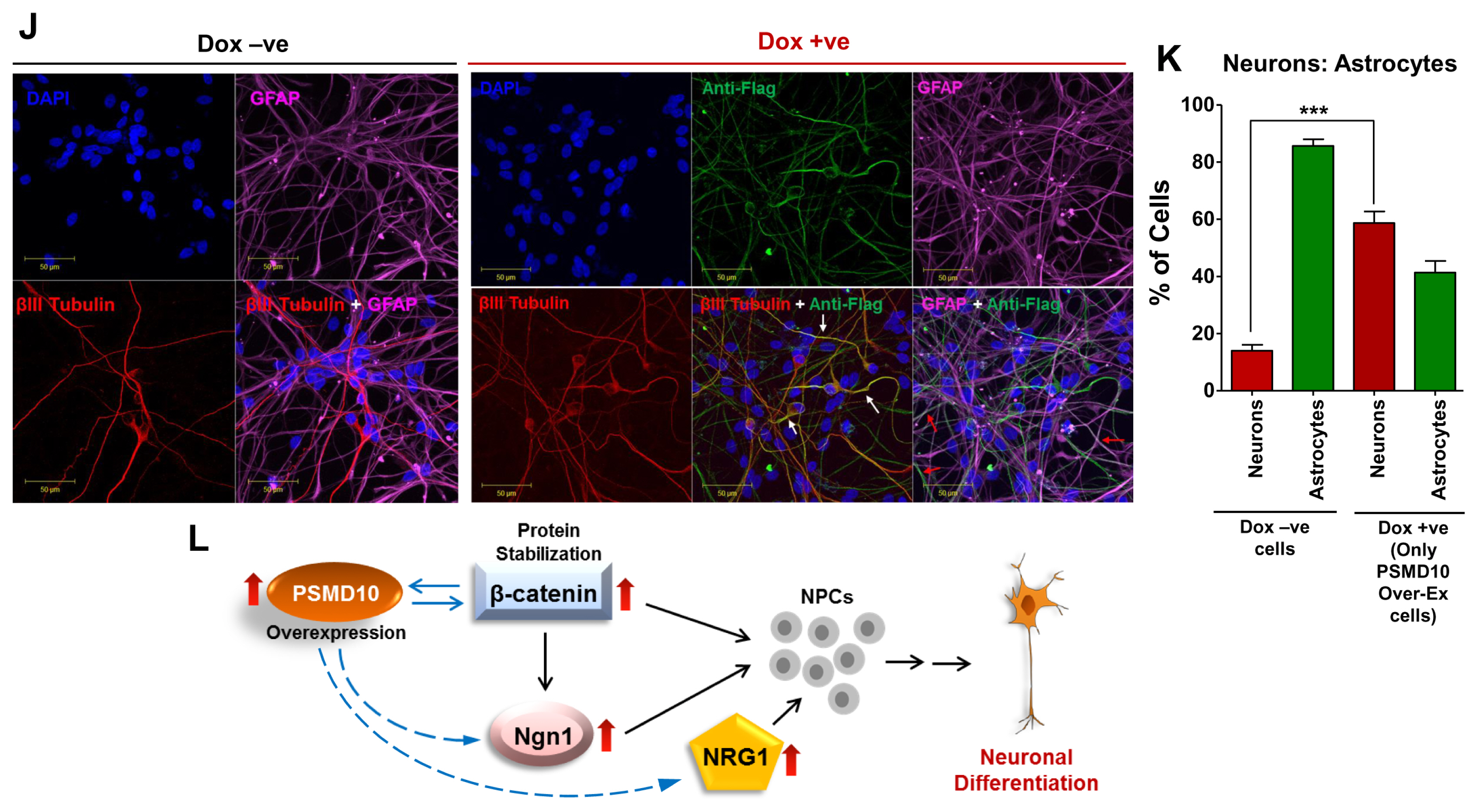
- PSMD10(Gankyrin), a proteasome assembly chaperone, is a widely known oncoprotein which aspects many hall mark properties of cancer. However, except proteasome assembly chaperon function its role in normal cell function remains unknown. To address this issue, we induced PSMD10(Gankyrin) overexpression in HEK293 cells and the resultant large-scale changes in gene expression profile were analyzed. We constituted networks from microarray data of these differentially expressed genes and carried out extensive topological analyses. The overrecurring yet consistent theme that appeared throughout analysis using varied network metrics is that all genes and interactions identified as important would be involved in neurogenesis and neuronal development. Intrigued we tested the possibility that PSMD10(Gankyrin) may be strongly associated with cell fate decisions that commit neural stem cells to differentiate into neurons. Overexpression of PSMD10(Gankyrin) in human neural progenitor cells facilitated neuronal differentiation via β-catenin Ngn1 pathway. Here for the first time we provide preliminary and yet compelling experimental evidence for the involvement of a potential oncoprotein - PSMD10(Gankyrin), in neuronal differentiation.
MeSH Terms
Figure
Reference
-
References
1. Hori T, Kato S, Saeki M, DeMartino GN, Slaughter CA, Takeuchi J, Toh-e A, Tanaka K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene. 1998; 216:113–122. DOI: 10.1016/S0378-1119(98)00309-6. PMID: 9714768.
Article2. Wang X, Jiang B, Zhang Y. Gankyrin regulates cell signaling network. Tumour Biol. 2016; 37:5675–5682. DOI: 10.1007/s13277-016-4854-z. PMID: 26819208.
Article3. Li H, Zhang J, Zhen C, Yang B, Feng L. Gankyrin as a potential target for tumor therapy: evidence and perspectives. Am J Transl Res. 2018; 10:1949–1960. PMID: 30093934. PMCID: PMC6079124.4. Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006; 45:15168–15178. DOI: 10.1021/bi062188q. PMID: 17176038.
Article5. Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004; 13:1435–1448. DOI: 10.1110/ps.03554604. PMID: 15152081. PMCID: PMC2279977.
Article6. Dawson S, Higashitsuji H, Wilkinson AJ, Fujita J, Mayer RJ. Gankyrin: a new oncoprotein and regulator of pRb and p53. Trends Cell Biol. 2006; 16:229–233. DOI: 10.1016/j.tcb.2006.03.001. PMID: 16581249.
Article7. Higashitsuji H, Liu Y, Mayer RJ, Fujita J. The oncoprotein gankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 2005; 4:1335–1337. DOI: 10.4161/cc.4.10.2107. PMID: 16177571.
Article8. Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010; 20:391–401. DOI: 10.1016/j.tcb.2010.03.007. PMID: 20427185. PMCID: PMC2902798.
Article9. Chen Y, Li HH, Fu J, Wang XF, Ren YB, Dong LW, Tang SH, Liu SQ, Wu MC, Wang HY. Oncoprotein p28 GANK binds to RelA and retains NF-kappaB in the cytoplasm through nuclear export. Cell Res. 2007; 17:1020–1029. DOI: 10.1038/cr.2007.99. PMID: 18040287.
Article10. Dong LW, Yang GZ, Pan YF, Chen Y, Tan YX, Dai RY, Ren YB, Fu J, Wang HY. The oncoprotein p28GANK establishes a positive feedback loop in β-catenin signaling. Cell Res. 2011; 21:1248–1261. DOI: 10.1038/cr.2011.103. PMID: 21691299. PMCID: PMC3193485.
Article11. Nanaware PP, Ramteke MP, Somavarapu AK, Venkatraman P. Discovery of multiple interacting partners of gankyrin, a proteasomal chaperone and an oncoprotein--evidence for a common hot spot site at the interface and its functional relevance. Proteins. 2014; 82:1283–1300. DOI: 10.1002/prot.24494. PMID: 24338975.
Article12. Banerjee SJ, Sinha S, Roy S. Slow poisoning and destruction of networks: edge proximity and its implications for biological and infrastructure networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2015; 91:022807. DOI: 10.1103/PhysRevE.91.022807. PMID: 25768552.
Article13. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550. DOI: 10.1073/pnas.0506580102. PMID: 16199517. PMCID: PMC1239896.
Article14. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57. DOI: 10.1038/nprot.2008.211. PMID: 19131956.
Article15. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. DOI: 10.1093/nar/gkn923. PMID: 19033363. PMCID: PMC2615629.
Article16. Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001; 104:365–376. DOI: 10.1016/S0092-8674(01)00224-0. PMID: 11239394.
Article17. Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004; 131:2791–2801. DOI: 10.1242/dev.01165. PMID: 15142975.
Article18. Sato T, Sato F, Kamezaki A, Sakaguchi K, Tanigome R, Kawakami K, Sehara-Fujisawa A. Neuregulin 1 type II-ErbB signaling promotes cell divisions generating neurons from neural progenitor cells in the developing zebrafish brain. PLoS One. 2015; 10:e0127360. DOI: 10.1371/journal.pone.0127360. PMID: 26001123. PMCID: PMC4441363.
Article19. Gambarotta G, Fregnan F, Gnavi S, Perroteau I. Neuregulin 1 role in Schwann cell regulation and potential applications to promote peripheral nerve regeneration. Int Rev Neurobiol. 2013; 108:223–256. DOI: 10.1016/B978-0-12-410499-0.00009-5. PMID: 24083437.
Article20. Mellodew K, Suhr R, Uwanogho DA, Reuter I, Lendahl U, Hodges H, Price J. Nestin expression is lost in a neural stem cell line through a mechanism involving the proteasome and Notch signalling. Brain Res Dev Brain Res. 2004; 151:13–23. DOI: 10.1016/j.devbrainres.2004.03.018. PMID: 15246688.
Article21. Sahu I, Sangith N, Ramteke M, Gadre R, Venkatraman P. A novel role for the proteasomal chaperone PSMD9 and hnRNPA1 in enhancing IκBα degradation and NF-κB activation - functional relevance of predicted PDZ domain-motif interaction. FEBS J. 2014; 281:2688–2709. DOI: 10.1111/febs.12814. PMID: 24720748.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Functional characterization of human oncoprotein gankyrin in Zebrafish
- Neural Stem Cells and Ischemic Brain
- Neurogenesis and Regulation of Olfactory Epithelium
- Adult Neurogenesis in the Central and Peripheral Nervous Systems
- Nestin expressing progenitor cells during establishment of the neural retina and its vasculature