Ann Dermatol.
2018 Aug;30(4):432-440. 10.5021/ad.2018.30.4.432.
Possible Role of Single Stranded DNA Binding Protein 3 on Skin Hydration by Regulating Epidermal Differentiation
- Affiliations
-
- 1Department of Dermatology, College of Medicine, Chungnam National University, Daejeon, Korea. ttole17@naver.com
- 2ichrogene, Suwon, Korea.
- 3LG Household and Healthcare, Daejeon, Korea.
- 4Department of Bio-Cosmetic Science, Seowon University, Cheongju, Korea.
- KMID: 2457520
- DOI: http://doi.org/10.5021/ad.2018.30.4.432
Abstract
- BACKGROUND
Skin hydration is a common problem both in elderly and young people as dry skin may cause irritation, dermatological disorders, and wrinkles. While both genetic and environmental factors seem to influence skin hydration, thorough genetic studies on skin hydration have not yet been conducted.
OBJECTIVE
We used a genome-wide association study (GWAS) to explore the genetic elements underlying skin hydration by regulating epidermal differentiation and skin barrier function.
METHODS
A GWAS was conducted to investigate the genetic factors influencing skin hydration in 100 Korean females along with molecular studies of genes in human epidermal keratinocytes for functional study in vitro.
RESULTS
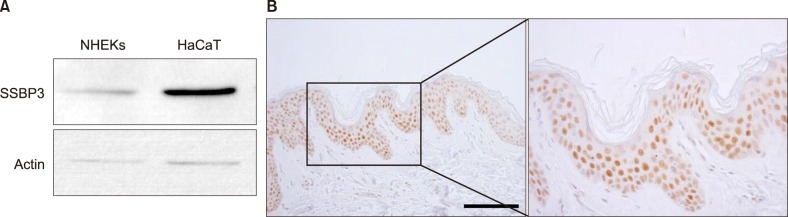
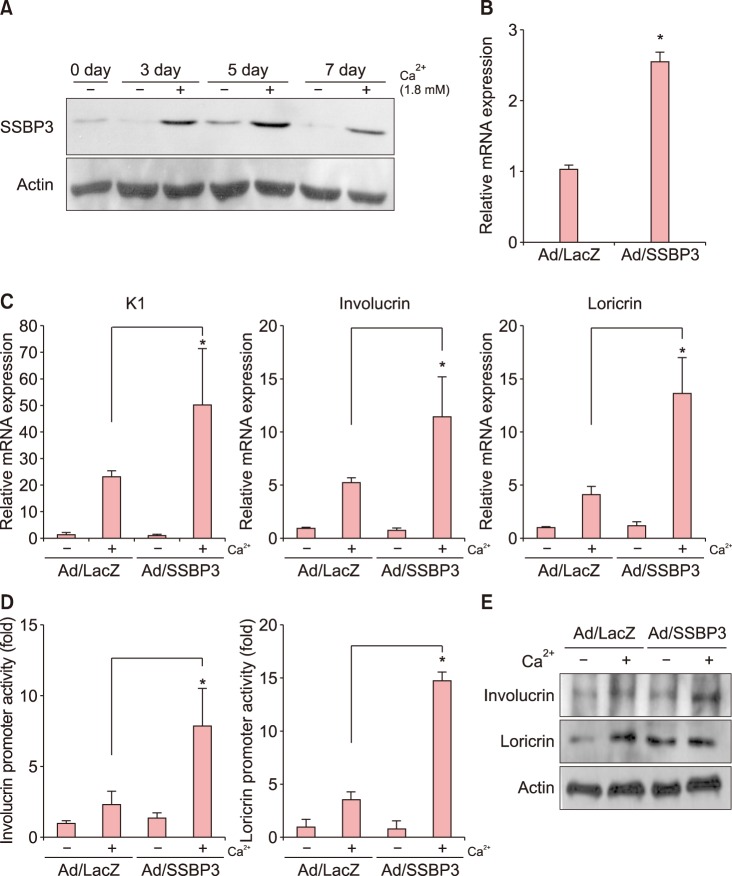
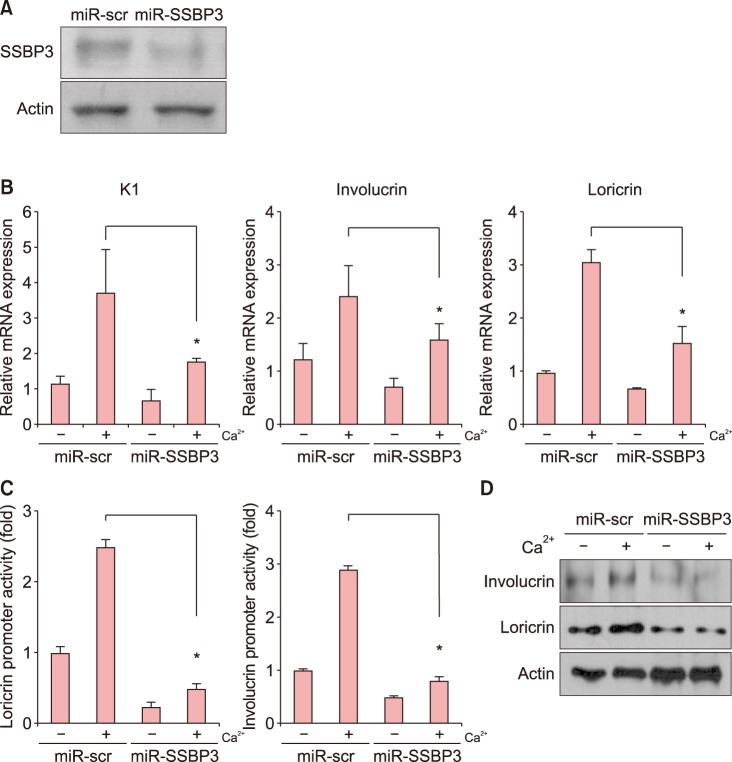
Among several single nucleotide polymorphisms identified in GWAS, we focused on Single Stranded DNA Binding Protein 3 (SSBP3) which is associated with DNA replication and DNA damage repair. To better understand the role of SSBP3 in skin cells, we introduced a calcium-induced differentiation keratinocyte culture system model and found that SSBP3 was upregulated in keratinocytes in a differentiation dependent manner. When SSBP3 was overexpressed using a recombinant adenovirus, the expression of differentiation-related genes such as loricrin and involucrin was markedly increased.
CONCLUSION
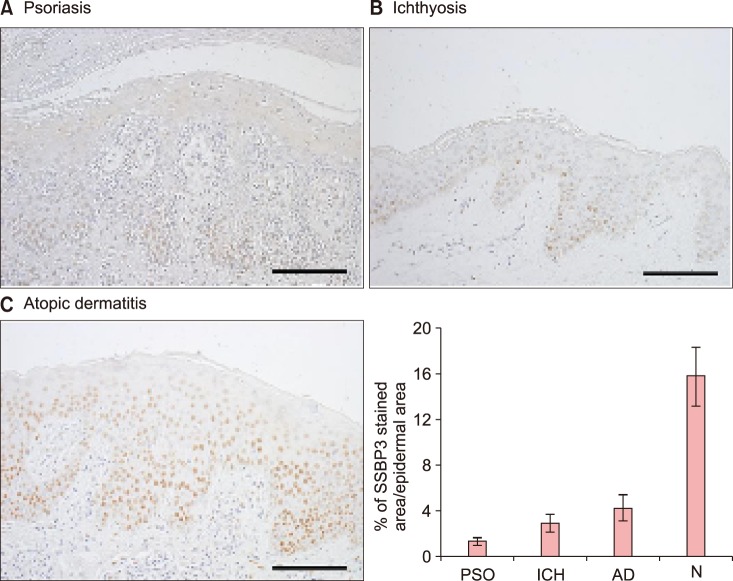
Taken together, our results suggest that genetic variants in the intronic region of SSBP3 could be determinants in skin hydration of Korean females. SSBP3 represents a new candidate gene to evaluate the molecular basis of the hydration ability in individuals.
Keyword
MeSH Terms
Figure
Reference
-
1. White-Chu EF, Reddy M. Dry skin in the elderly: complexities of a common problem. Clin Dermatol. 2011; 29:37–42. PMID: 21146730.
Article2. Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. Bioessays. 2002; 24:789–800. PMID: 12210515.
Article3. Lebwohl M, Herrmann LG. Impaired skin barrier function in dermatologic disease and repair with moisturization. Cutis. 2005; 76(6 Suppl):7–12. PMID: 16869176.4. Candille SI, Absher DM, Beleza S, Bauchet M, McEvoy B, Garrison NA, et al. Genome-wide association studies of quantitatively measured skin, hair, and eye pigmentation in four European populations. PLoS One. 2012; 7:e48294. PMID: 23118974.
Article5. Zhang M, Song F, Liang L, Nan H, Zhang J, Liu H, et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum Mol Genet. 2013; 22:2948–2959. PMID: 23548203.
Article6. Nan H, Kraft P, Qureshi AA, Guo Q, Chen C, Hankinson SE, et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J Invest Dermatol. 2009; 129:2250–2257. PMID: 19340012.
Article7. Lee JS, Kim DH, Choi DK, Kim CD, Ahn GB, Yoon TY, et al. Comparison of gene expression profiles between keratinocytes, melanocytes and fibroblasts. Ann Dermatol. 2013; 25:36–45. PMID: 23467683.
Article8. Shi G, Sohn KC, Kim SY, Ryu EK, Park YS, Lee Y, et al. Sox9 increases the proliferation and colony-forming activity of outer root sheath cells cultured in vitro. Ann Dermatol. 2011; 23:138–143. PMID: 21747610.9. Kalia YN, Nonato LB, Lund CH, Guy RH. Development of skin barrier function in premature infants. J Invest Dermatol. 1998; 111:320–326. PMID: 9699737.
Article10. Sevilla LM, Nachat R, Groot KR, Klement JF, Uitto J, Djian P, et al. Mice deficient in involucrin, envoplakin, and periplakin have a defective epidermal barrier. J Cell Biol. 2007; 179:1599–1612. PMID: 18166659.
Article11. Engelke M, Jensen JM, Ekanayake-Mudiyanselage S, Proksch E. Effects of xerosis and ageing on epidermal proliferation and differentiation. Br J Dermatol. 1997; 137:219–225. PMID: 9292070.
Article12. Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008; 4:e1000074. PMID: 18483556.
Article13. Rees JL. The genetics of sun sensitivity in humans. Am J Hum Genet. 2004; 75:739–751. PMID: 15372380.
Article14. Kwak TJ, Chang YH, Shin YA, Shin JM, Kim JH, Lim SK, et al. Identification of a possible susceptibility locus for UVB-induced skin tanning phenotype in Korean females using genomewide association study. Exp Dermatol. 2015; 24:942–946. PMID: 26174610.
Article15. Ashton NW, Bolderson E, Cubeddu L, O'Byrne KJ, Richard DJ. Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BMC Mol Biol. 2013; 14:9. PMID: 23548139.
Article16. Castro P, Liang H, Liang JC, Nagarajan L. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics. 2002; 80:78–85. PMID: 12079286.
Article17. Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, et al. Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc Natl Acad Sci U S A. 2002; 99:14320–14325. PMID: 12381786.
Article18. van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003; 130:1915–1925. PMID: 12642495.
Article19. Nishioka N, Nagano S, Nakayama R, Kiyonari H, Ijiri T, Taniguchi K, et al. Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1-Ldb1 complex. Development. 2005; 132:2535–2546. PMID: 15857913.
Article20. Enkhmandakh B, Makeyev AV, Bayarsaihan D. The role of the proline-rich domain of Ssdp1 in the modular architecture of the vertebrate head organizer. Proc Natl Acad Sci U S A. 2006; 103:11631–11636. PMID: 16864769.
Article21. Cai Y, Xu Z, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance and function of the LIM-homeodomain transcription factor LHX2 in pituitary cells. Biochem Biophys Res Commun. 2008; 373:303–308. PMID: 18565323.
Article22. Liu J, Luo X, Xu Y, Gu J, Tang F, Jin Y, et al. Single-stranded DNA binding protein Ssbp3 induces differentiation of mouse embryonic stem cells into trophoblast-like cells. Stem Cell Res Ther. 2016; 7:79. PMID: 27236334.
Article23. Moabbi AM, Agarwal N, El Kaderi B, Ansari A. Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A. 2012; 109:8505–8510. PMID: 22586116.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Erratum: Possible Role of Single Stranded DNA Binding Protein 3 on Skin Hydration by Regulating Epidermal Differentiation
- Nuclear Factors Bind to the Specific Sites in Promoter Region of Human YB-1 Gene
- The binding of ethidium bromide with DNA: interaction with single- and double-stranded structures
- Compiling Multicopy Single-Stranded DNA Sequences from Bacterial Genome Sequences
- Rab25 Deficiency Perturbs Epidermal Differentiation and Skin Barrier Function in Mice