Cancer Res Treat.
2019 Jul;51(3):1198-1206. 10.4143/crt.2018.527.
Associations of Genetic Variations in Mismatch Repair Genes MSH3 and PMS1 with Acute Adverse Events and Survival in Patients with Rectal Cancer Receiving Postoperative Chemoradiotherapy
- Affiliations
-
- 1State Key Laboratory of Molecular Oncology, Department of Etiology & Carcinogenesis, Beijing Key Laboratory for Carcinogenesis and Cancer Prevention, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China. tanwen68@hotmail.com
- 2Department of Radiation Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China. jingjin1025@163.com
- KMID: 2454311
- DOI: http://doi.org/10.4143/crt.2018.527
Abstract
- PURPOSE
Mismatch repair (MMR) deficiency plays a critical role in rectal cancer. This study aimed to explore the associations between genetic variations in seven MMR genes and adverse events (AEs) and survival of patients with rectal cancer treated with postoperative chemoradiotherapy (CRT).
MATERIALS AND METHODS
Fifty single nucleotide polymorphisms in seven MMR (MLH1, MLH3, MSH2, MSH3, MSH6, PMS1 and PMS2) genes were genotyped by Sequenom MassARRAY method in 365 patients with locally advanced rectal cancer receiving postoperative CRT. The associations between genotypes and AEs were measured by odds ratios and 95% confidence intervals (CIs) by unconditional logistic regression model. The associations between genetic variations and survival were computed by the hazard ratios and 95% CIs by Cox proportional regression model.
RESULTS
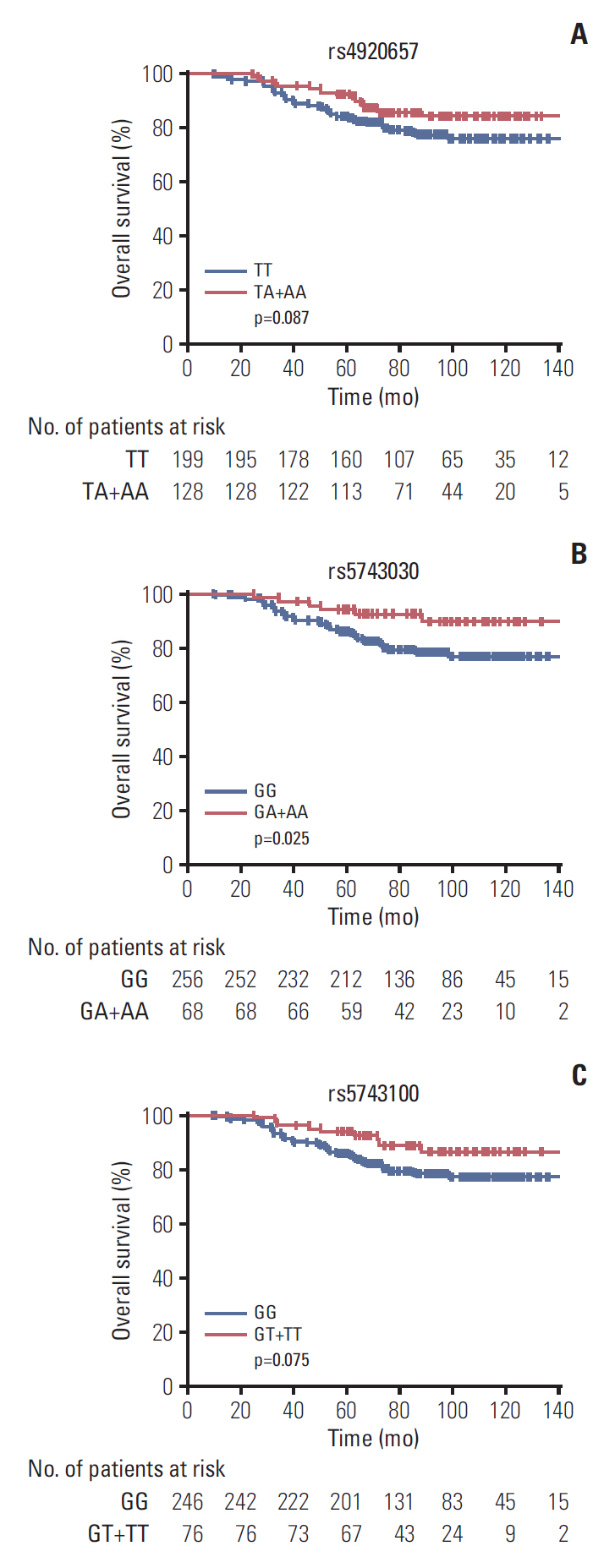
The most common grade ≥ 2 AEs in those 365 patients, in decreasing order, were diarrhea (44.1%), leukopenia (29.6%), and dermatitis (18.9%). Except 38 cases missing, 61 patients (18.7%) died during the follow-up period. We found MSH3 rs12513549, rs33013 and rs6151627 significantly associated with the risk of grade ≥ 2 diarrhea. PMS1 rs1233255 had an impact on the occurrence of grade ≥2 dermatitis. Meanwhile, PMS1 rs4920657, rs5743030, and rs5743100 were associated with overall survival (OS) time of rectal cancer.
CONCLUSION
These results suggest that MSH3 and PMS1 polymorphisms may play important roles in AEs prediction and prognosis of rectal cancer patients receiving postoperative CRT, which can be potential genetic biomarkers for rectal cancer personalized treatment.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016; 66:115–32.
Article2. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010; 375:1030–47.
Article3. Sauer R, Becker H, Hohenberger W, Rodel C, Wittekind C, Fietkau R, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004; 351:1731–40.
Article4. Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat Rev Cancer. 2003; 3:912–20.
Article5. Deenen MJ, Tol J, Burylo AM, Doodeman VD, de Boer A, Vincent A, et al. Relationship between single nucleotide polymorphisms and haplotypes in DPYD and toxicity and efficacy of capecitabine in advanced colorectal cancer. Clin Cancer Res. 2011; 17:3455–68.
Article6. Huang Y, Feng Y, Ren H, Zhang M, Li H, Qiao Y, et al. Associations of genetic variations in microRNA seed regions with acute adverse events and survival in patients with rectal cancer receiving postoperative chemoradiation therapy. Int J Radiat Oncol Biol Phys. 2018; 100:1026–33.
Article7. Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013; 5:a012633.
Article8. O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR system. Carcinogenesis. 2006; 27:682–92.9. de la Chapelle A. Microsatellite instability. N Engl J Med. 2003; 349:209–10.
Article10. de Rosa N, Rodriguez-Bigas MA, Chang GJ, Veerapong J, Borras E, Krishnan S, et al. DNA mismatch repair deficiency in rectal cancer: benchmarking its impact on prognosis, neoadjuvant response prediction, and clinical cancer genetics. J Clin Oncol. 2016; 34:3039–46.
Article11. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–20.12. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017; 357:409–13.13. Liu JY, Qian CY, Gao YF, Chen J, Zhou HH, Yin JY. Association between DNA mismatch repair gene polymorphisms and platinum-based chemotherapy toxicity in non-small cell lung cancer patients. Chin J Cancer. 2017; 36:12.
Article14. Vymetalkova V, Pardini B, Rosa F, Di Gaetano C, Novotny J, Levy M, et al. Variations in mismatch repair genes and colorectal cancer risk and clinical outcome. Mutagenesis. 2014; 29:259–65.
Article15. Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006; 106:302–23.
Article16. Kumar C, Williams GM, Havens B, Dinicola MK, Surtees JA. Distinct requirements within the Msh3 nucleotide binding pocket for mismatch and double-strand break repair. J Mol Biol. 2013; 425:1881–98.
Article17. van Oers JM, Edwards Y, Chahwan R, Zhang W, Smith C, Pechuan X, et al. The MutSbeta complex is a modulator of p53-driven tumorigenesis through its functions in both DNA double-strand break repair and mismatch repair. Oncogene. 2014; 33:3939–46.18. Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D. High resolution deletion mapping reveals frequent allelic losses at the DNA mismatch repair loci hMLH1 and hMSH3 in non-small cell lung cancer. Int J Cancer. 1998; 77:173–80.19. Kawakami T, Shiina H, Igawa M, Deguchi M, Nakajima K, Ogishima T, et al. Inactivation of the hMSH3 mismatch repair gene in bladder cancer. Biochem Biophys Res Commun. 2004; 325:934–42.
Article20. Tseng-Rogenski SS, Hamaya Y, Choi DY, Carethers JM. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology. 2015; 148:579–89.
Article21. Berndt SI, Platz EA, Fallin MD, Thuita LW, Hoffman SC, Helzlsouer KJ. Mismatch repair polymorphisms and the risk of colorectal cancer. Int J Cancer. 2007; 120:1548–54.
Article22. Koessler T, Azzato EM, Perkins B, Macinnis RJ, Greenberg D, Easton DF, et al. Common germline variation in mismatch repair genes and survival after a diagnosis of colorectal cancer. Int J Cancer. 2009; 124:1887–91.
Article23. Larrea AA, Lujan SA, Kunkel TA. SnapShot: DNA mismatch repair. Cell. 2010; 141:730.e1.
Article24. Gatzidou E, Michailidi C, Tseleni-Balafouta S, Theocharis S. An epitome of DNA repair related genes and mechanisms in thyroid carcinoma. Cancer Lett. 2010; 290:139–47.
Article25. Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005; 4:227–32.26. Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003; 349:247–57.
Article27. Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007; 133:1858–68.
Article28. Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998; 4:1–6.29. Dong J, Hu Z, Shu Y, Pan S, Chen W, Wang Y, et al. Potentially functional polymorphisms in DNA repair genes and nonsmall-cell lung cancer survival: a pathway-based analysis. Mol Carcinog. 2012; 51:546–52.
Article30. Yang J, Wang X, Zou SM, Li HM, Xiao Q, Feng YR, et al. Genetic variations in MLH3 and MSH2 genes are associated with the sensitivity and prognosis in locally advanced rectal cancer patients receiving preoperative chemoradiotherapy. Zhonghua Zhong Liu Za Zhi. 2018; 40:433–40.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Hereditary Nonpolyposis Colorectal Cancer
- Neoadjuvant chemoradiotherapy determines the prognostic impact of anastomotic leakage in advanced rectal cancer
- Expression Pattern of DNA Mismatch Repair Genes in Tumors of Microsatellite Mutator Phenotype
- Gastric Cancer and DNA Mismatch Repair
- Role of Radiation Therapy as an Adjuvant Treatment in Rectal Cancer Management