J Breast Cancer.
2017 Sep;20(3):286-296. 10.4048/jbc.2017.20.3.286.
NanoString nCounter® Approach in Breast Cancer: A Comparative Analysis with Quantitative Real-Time Polymerase Chain Reaction, In Situ Hybridization, and Immunohistochemistry
- Affiliations
-
- 1Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. eunyoon.cho@samsung.com
- 2Department of Pathology, Gangnam Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea.
- 3Department of Pathology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 4Division of Hematology-Oncology, Department of Internal Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2438996
- DOI: http://doi.org/10.4048/jbc.2017.20.3.286
Abstract
- PURPOSE
Accurate testing for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) is essential for breast cancer treatment. At present, immunohistochemistry (IHC)/florescence in situ hybridization (FISH) are widely accepted as the standard testing methods. To investigate the value of NanoString nCounter®, we performed its comparative analysis with IHC/FISH and real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) for the assessment of ER, PR, and HER2.
METHODS
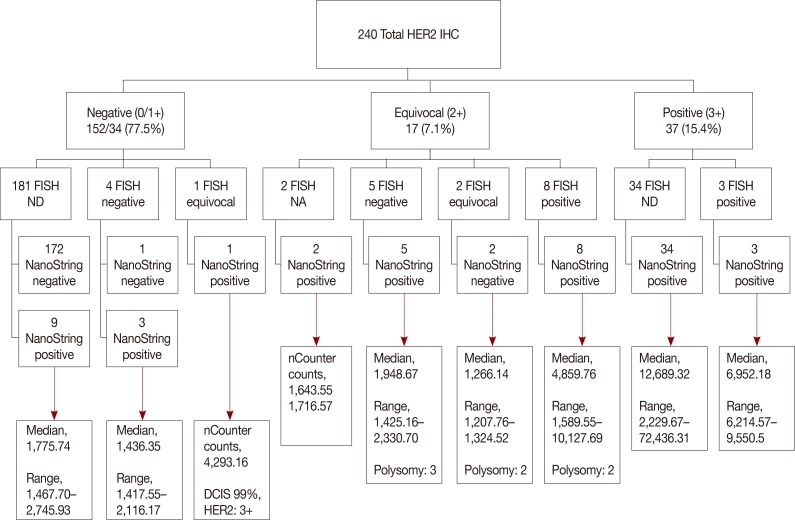
Data on IHC/FISH results for ER, PR, and HER2 in 240 patients from a single tertiary hospital in Korea were collected and compared with NanoString nCounter® and qRT-PCR results at a single institution.
RESULTS
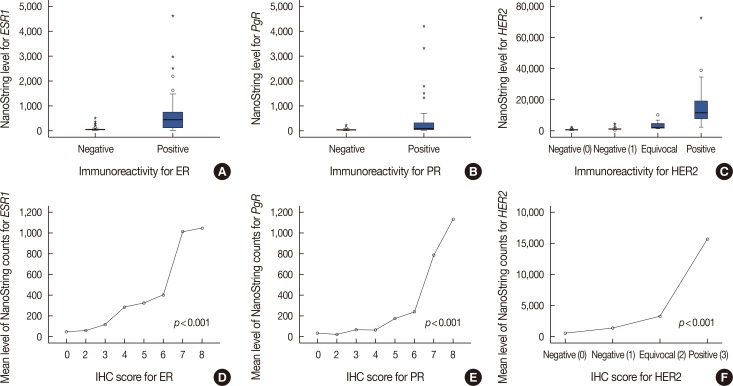
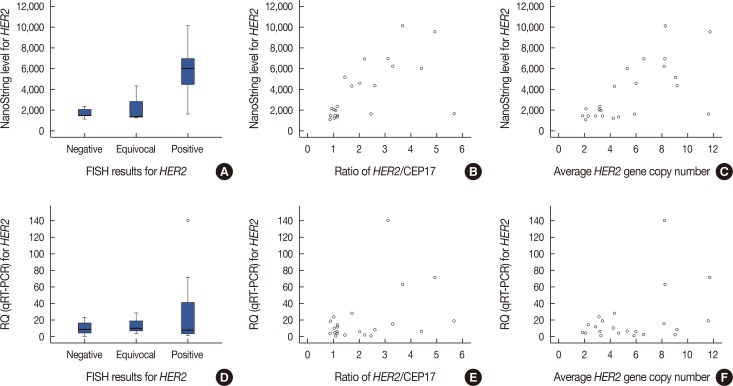
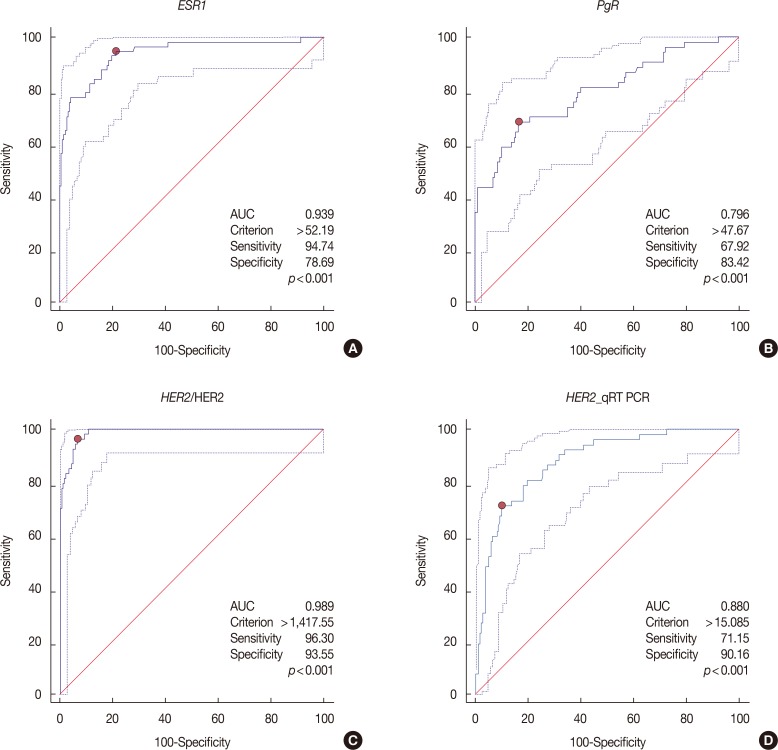
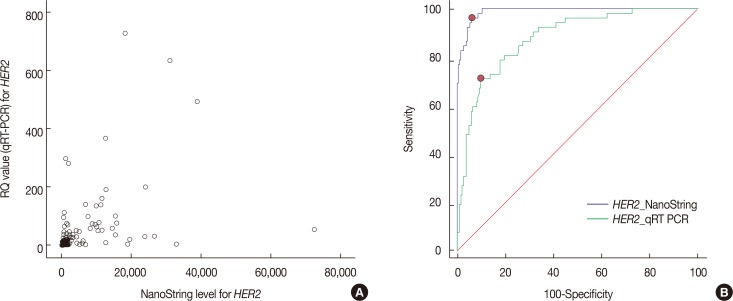
Expression levels for each gene using NanoString nCounter® showed good correlation with the corresponding data for protein expression by IHC (p<0.001) and gene amplification status for HER2 (p<0.001). Comparisons between gene expression and IHC data showed good overall agreement with a high area under the curve (AUC) for ESR1/ER (AUC=0.939), PgR/PR (AUC=0.796), and HER2/HER2 (AUC=0.989) (p<0.001).
CONCLUSION
The quantification of ER, PgR, and HER2 mRNA expression with NanoString nCounter® may be a viable alternative to conventional IHC/FISH methods.
MeSH Terms
-
Breast Neoplasms*
Breast*
Estrogens
Gene Amplification
Gene Expression
Humans
Immunohistochemistry*
In Situ Hybridization*
Korea
Polymerase Chain Reaction
Real-Time Polymerase Chain Reaction*
Receptor, Epidermal Growth Factor
Receptors, Progesterone
Reverse Transcription
RNA, Messenger
Tertiary Care Centers
Estrogens
RNA, Messenger
Receptor, Epidermal Growth Factor
Receptors, Progesterone
Figure
Reference
-
1. Dowsett M, Houghton J, Iden C, Salter J, Farndon J, A'Hern R, et al. Benefit from adjuvant tamoxifen therapy in primary breast cancer patients according oestrogen receptor, progesterone receptor, EGF receptor and HER2 status. Ann Oncol. 2006; 17:818–826. PMID: 16497822.
Article2. Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999; 17:1474–1481. PMID: 10334533.
Article3. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010; 28:2784–2795. PMID: 20404251.
Article4. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987; 235:177–182. PMID: 3798106.5. Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004; 5:63–69. PMID: 15140287.
Article6. Montemurro F, Scaltriti M. Biomarkers of drugs targeting HER-family signaling in cancer. J Pathol. 2014; 232:219–229. PMID: 24105684.7. Tandon AK, Clark GM, Chamness GC, Ullrich A, McGuire WL. HER-2/neu oncogene protein and prognosis in breast cancer. J Clin Oncol. 1989; 7:1120–1128. PMID: 2569032.
Article8. Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002; 20:719–726. PMID: 11821453.
Article9. Marty M, Cognetti F, Maraninchi D, Snyder R, Mauriac L, Tubiana-Hulin M, et al. Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as first-line treatment: the M77001 study group. J Clin Oncol. 2005; 23:4265–4274. PMID: 15911866.
Article10. Joensuu H, Kellokumpu-Lehtinen PL, Bono P, Alanko T, Kataja V, Asola R, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med. 2006; 354:809–820. PMID: 16495393.
Article11. Montemurro F, Valabrega G, Aglietta M. Lapatinib: a dual inhibitor of EGFR and HER2 tyrosine kinase activity. Expert Opin Biol Ther. 2007; 7:257–268. PMID: 17250463.
Article12. Baselga J, Cortés J, Kim SB, Im SA, Hegg R, Im YH, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012; 366:109–119. PMID: 22149875.
Article13. Banerjee S, Smith IE. Management of small HER2-positive breast cancers. Lancet Oncol. 2010; 11:1193–1199. PMID: 21126688.
Article14. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: breast cancer 2013. Accessed August 18th, 2017. https://www.nccn.org/.15. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013; 31:3997–4013. PMID: 24101045.
Article16. Hanna WM, Rüschoff J, Bilous M, Coudry RA, Dowsett M, Osamura RY, et al. HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014; 27:4–18.
Article17. Laudadio J, Quigley DI, Tubbs R, Wolff DJ. HER2 testing: a review of detection methodologies and their clinical performance. Expert Rev Mol Diagn. 2007; 7:53–64. PMID: 17187484.
Article18. Lehmann-Che J, Amira-Bouhidel F, Turpin E, Antoine M, Soliman H, Legres L, et al. Immunohistochemical and molecular analyses of HER2 status in breast cancers are highly concordant and complementary approaches. Br J Cancer. 2011; 104:1739–1746. PMID: 21540864.
Article19. Barberis M, Pellegrini C, Cannone M, Arizzi C, Coggi G, Bosari S. Quantitative PCR and HER2 testing in breast cancer: a technical and cost-effectiveness analysis. Am J Clin Pathol. 2008; 129:563–570. PMID: 18343783.20. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008; 26:317–325. PMID: 18278033.
Article21. Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998; 11:155–168. PMID: 9504686.22. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752. PMID: 10963602.
Article23. Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010; 23(Suppl 2):S60–S64. PMID: 20436504.
Article24. Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014; 14:177. PMID: 24625003.
Article25. Lira ME, Choi YL, Lim SM, Deng S, Huang D, Ozeck M, et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn. 2014; 16:229–243. PMID: 24418728.
Article26. Watters AD, Going JJ, Cooke TG, Bartlett JM. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003; 77:109–114. PMID: 12602909.
Article27. Hyun CL, Lee HE, Kim KS, Kim SW, Kim JH, Choe G, et al. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol. 2008; 61:317–321. PMID: 17761736.
Article28. Hofmann M, Stoss O, Gaiser T, Kneitz H, Heinmöller P, Gutjahr T, et al. Central HER2 IHC and FISH analysis in a trastuzumab (Herceptin) phase II monotherapy study: assessment of test sensitivity and impact of chromosome 17 polysomy. J Clin Pathol. 2008; 61:89–94. PMID: 17412870.
Article29. Seidman AD, Berry D, Cirrincione C, Harris L, Muss H, Marcom PK, et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: final results of Cancer and Leukemia Group B protocol 9840. J Clin Oncol. 2008; 26:1642–1649. PMID: 18375893.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Amplification of the UQCRFS1 Gene in Gastric Cancers
- Detection of HPV in tissue of cervical lesion: Comparative study between in situ hybridization and PCR in situ hybridization
- Expression of vascular endothelial growth factor in oral squamous cell carcinoma
- Evaluation of HER2/neu Status by Real-Time Quantitative PCR in Breast Cancer
- High MicroRNA-370 Expression Correlates with Tumor Progression and Poor Prognosis in Breast Cancer