J Breast Cancer.
2015 Dec;18(4):323-328. 10.4048/jbc.2015.18.4.323.
High MicroRNA-370 Expression Correlates with Tumor Progression and Poor Prognosis in Breast Cancer
- Affiliations
-
- 1Department of Pathology, Hanyang University College of Medicine, Seoul, Korea. medartisan@hanyang.ac.kr
- 2Department of Surgery, Hanyang University College of Medicine, Seoul, Korea.
- KMID: 2176280
- DOI: http://doi.org/10.4048/jbc.2015.18.4.323
Abstract
- PURPOSE
Deregulation of microRNA-370 (miR-370) has been reported in various cancers, in which it can act as either an oncogene or a tumor suppressor gene. However, the clinicopathologic significance of miR-370 expression in breast cancer has not been studied.
METHODS
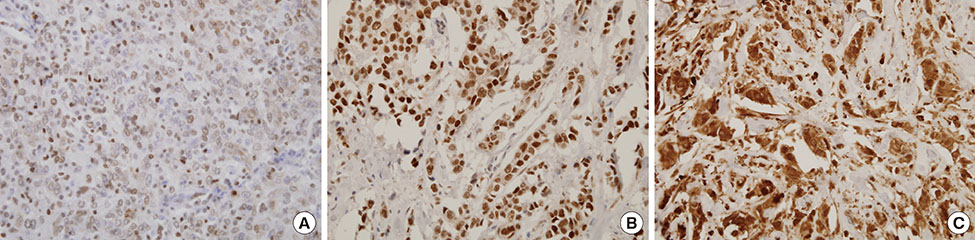
The expression of miR-370 was determined with quantitative real-time polymerase chain reaction in 60 formalin-fixed, paraffin-embedded primary breast cancer tissues. Additionally, the protein expression levels of previously known targets of miR-370, such as FOXM1, FOXO1, and FOXO3a, were detected using immunohistochemistry. Finally, we analyzed its correlation with target protein expression, clinicopathologic features, and clinical outcome.
RESULTS
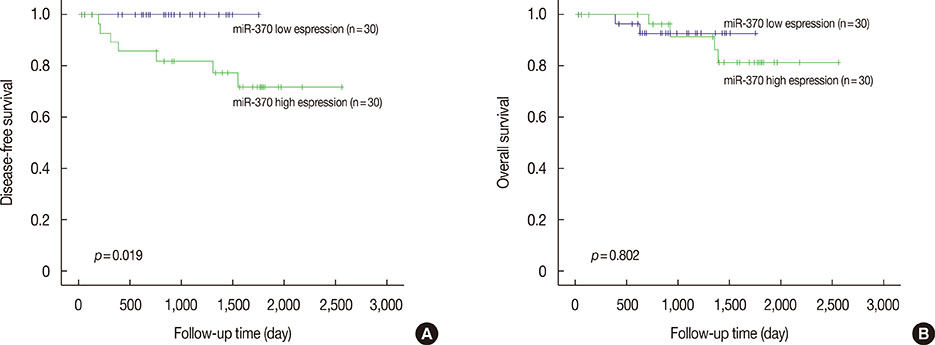
High levels of miR-370 expression correlated with lymph node metastasis (p=0.009), advanced stage (p=0.002), and frequent perineural invasion (p=0.042). Moreover, patients with high miR-370 expression had poor disease-free survival compared with the low-expression group. However, no correlation was observed between miR-370 and its target protein expression.
CONCLUSION
Our results indicate that upregulation of miR-370 in breast cancer is correlated with breast cancer progression and that it might be a potential biomarker for predicting clinical outcomes.
MeSH Terms
Figure
Reference
-
1. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233.
Article2. Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007; 6:2127–2132.
Article3. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005; 65:7065–7070.
Article4. Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007; 8:R214.
Article5. Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009; 11:R27.6. Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008; 105:13021–13026.
Article7. Rothé F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011; 6:e20980.
Article8. Wu Z, Sun H, Zeng W, He J, Mao X. Upregulation of MircoRNA-370 induces proliferation in human prostate cancer cells by downregulating the transcription factor FOXO1. PLoS One. 2012; 7:e45825.
Article9. Cao X, Liu D, Yan X, Zhang Y, Yuan L, Zhang T, et al. Stat3 inhibits WTX expression through up-regulation of microRNA-370 in Wilms tumor. FEBS Lett. 2013; 587:639–644.
Article10. Fan C, Liu S, Zhao Y, Han Y, Yang L, Tao G, et al. Upregulation of miR-370 contributes to the progression of gastric carcinoma via suppression of FOXO1. Biomed Pharmacother. 2013; 67:521–526.
Article11. Lo SS, Hung PS, Chen JH, Tu HF, Fang WL, Chen CY, et al. Overexpression of miR-370 and downregulation of its novel target TGFβ-RII contribute to the progression of gastric carcinoma. Oncogene. 2012; 31:226–237.
Article12. Garcća-Ortć L, Cristóbal I, Cirauqui C, Guruceaga E, Marcotegui N, Calasanz MJ, et al. Integration of SNP and mRNA arrays with microRNA profiling reveals that MiR-370 is upregulated and targets NF1 in acute myeloid leukemia. PLoS One. 2012; 7:e47717.
Article13. Yungang W, Xiaoyu L, Pang T, Wenming L, Pan X. miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother. 2014; 68:149–154.
Article14. Feng Y, Wang L, Zeng J, Shen L, Liang X, Yu H, et al. FoxM1 is overexpressed in Helicobacter pylori-induced gastric carcinogenesis and is negatively regulated by miR-370. Mol Cancer Res. 2013; 11:834–844.
Article15. Zhang X, Zeng J, Zhou M, Li B, Zhang Y, Huang T, et al. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer. 2012; 11:56.
Article16. Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012; 18:BR299–BR308.
Article17. Xu WP, Yi M, Li QQ, Zhou WP, Cong WM, Yang Y, et al. Perturbation of MicroRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma. Hepatology. 2013; 58:1977–1991.
Article18. Chang KW, Chu TH, Gong NR, Chiang WF, Yang CC, Liu CJ, et al. miR-370 modulates insulin receptor substrate-1 expression and inhibits the tumor phenotypes of oral carcinoma. Oral Dis. 2013; 19:611–619.
Article19. Zhou M, Zeng J, Wang X, Guo Q, Huang T, Shen H, et al. MiR-370 sensitizes chronic myeloid leukemia K562 cells to homoharringtonine by targeting Forkhead box M1. J Transl Med. 2013; 11:265.
Article20. Jiang L, Cao XC, Cao JG, Liu F, Quan MF, Sheng XF, et al. Casticin induces ovarian cancer cell apoptosis by repressing FoxM1 through the activation of FOXO3a. Oncol Lett. 2013; 5:1605–1610.
Article21. Karadedou CT, Gomes AR, Chen J, Petkovic M, Ho KK, Zwolinska AK, et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2012; 31:1845–1858.
Article22. Aguirre-Gamboa R, Trevino V. SurvMicro: assessment of miRNA-based prognostic signatures for cancer clinical outcomes by multivariate survival analysis. Bioinformatics. 2014; 30:1630–1632.
Article23. Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014; 33:3014–3023.
Article24. Zhang X, Chen J, Radcliffe T, Lebrun DP, Tron VA, Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples. J Mol Diagn. 2008; 10:513–519.
Article25. Liu A, Xu X. MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol Biol. 2011; 724:259–267.
Article26. Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007; 7:36.
Article27. Hall JS, Taylor J, Valentine HR, Irlam JJ, Eustace A, Hoskin PJ, et al. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br J Cancer. 2012; 107:684–694.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast
- MicroRNA-222 Expression as a Predictive Marker for Tumor Progression in Hormone Receptor-Positive Breast Cancer
- The Long Noncoding RNA DUXAP8 Facilitates the Malignant Progression of Colon Cancer via the microRNA-378a-3p/FOXQ1 Axis
- MicroRNA-370 Regulates Cellepithelial-Mesenchymal Transition, Migration, Invasion, and Prognosis of Hepatocellular Carcinoma by Targeting GUCD1
- Quantitative Measurement of Serum MicroRNA-21 Expression in Relation to Breast Cancer Metastasis in Chinese Females