J Breast Cancer.
2017 Sep;20(3):240-245. 10.4048/jbc.2017.20.3.240.
Gene Regulatory Network Analysis for Triple-Negative Breast Neoplasms by Using Gene Expression Data
- Affiliations
-
- 1Department of Internal Medicine, Eulji University College of Medicine, Seoul, Korea.
- 2Department of Statistics, Keimyung University, Daegu, Korea.
- 3Department of Biomedical Sciences, Seoul National University College of Medicine, Seoul, Korea.
- 4Division of Fusion Data Analytics Laboratory, School of Industrial Management Engineering, Korea University, Seoul, Korea. swhan@korea.ac.kr
- KMID: 2438990
- DOI: http://doi.org/10.4048/jbc.2017.20.3.240
Abstract
- PURPOSE
To better identify the physiology of triple-negative breast neoplasm (TNBN), we analyzed the TNBN gene regulatory network using gene expression data.
METHODS
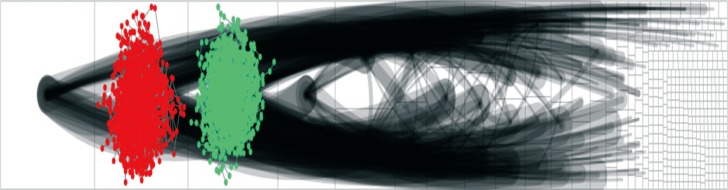
We collected TNBN gene expression data from The Cancer Genome Atlas to construct a TNBN gene regulatory network using least absolute shrinkage and selection operator regression. In addition, we constructed a triple-positive breast neoplasm (TPBN) network for comparison. Furthermore, survival analysis based on gene expression levels and differentially expressed gene (DEG) analysis were carried out to support and compare the network analysis results, respectively.
RESULTS
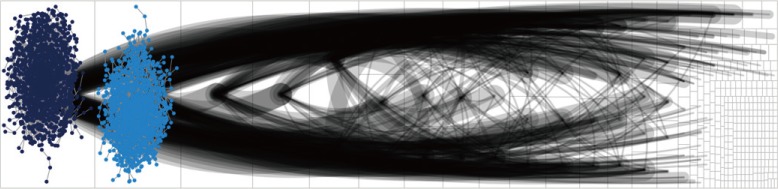
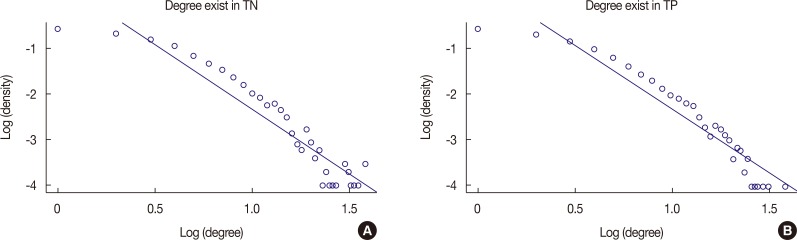
The TNBN gene regulatory network, which followed a power-law distribution, had 10,237 vertices and 17,773 edges, with an average vertex-to-vertex distance of 8.6. The genes ZDHHC20 and RAPGEF6 were identified by centrality analysis to be important vertices. However, in the DEG analysis, we could not find meaningful fold changes in ZDHHC20 and RAPGEF6 between the TPBN and TNBN gene expression data. In the multivariate survival analysis, the hazard ratio for ZDHHC20 and RAPGEF6 was 1.677 (1.192-2.357) and 1.676 (1.222-2.299), respectively.
CONCLUSION
Our TNBN gene regulatory network was a scale-free one, which means that the network would be easily destroyed if the hub vertices were attacked. Thus, it is important to identify the hub vertices in the network analysis. In the TNBN gene regulatory network, ZDHHC20 and RAPGEF6 were found to be oncogenes. Further study of these genes could help to reveal a novel method for treating TNBN in the future.
Keyword
MeSH Terms
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30. PMID: 26742998.
Article2. Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010; 7:e1000279. PMID: 20520800.
Article3. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–1948. PMID: 21067385.
Article4. Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, et al. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med. 2007; 357:2666–2676. PMID: 18160686.
Article5. de Matos Simoes R, Emmert-Streib F. Bagging statistical network inference from large-scale gene expression data. PLoS One. 2012; 7:e33624. PMID: 22479422.
Article6. Emmert-Streib F, de Matos Simoes R, Mullan P, Haibe-Kains B, Dehmer M. The gene regulatory network for breast cancer: integrated regulatory landscape of cancer hallmarks. Front Genet. 2014; 5:15. PMID: 24550935.
Article7. Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013; 45:1113–1120. PMID: 24071849.
Article8. Han SW, Chen G, Cheon MS, Zhong H. Estimation of directed acyclic graphs through two-stage adaptive lasso for gene network inference. J Am Stat Assoc. 2016; 111:1004–1019. PMID: 28239216.
Article9. Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the Lasso. Ann Stat. 2006; 34:1436–1462.
Article10. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. PMID: 15461798.11. Smyth GK. Limma: linear models for microarray data. In : Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer;2005. p. 397–420.12. Draper JM, Smith CD. DHHC20: a human palmitoyl acyltransferase that causes cellular transformation. Mol Membr Biol. 2010; 27:123–136. PMID: 20334580.
Article13. Gao X, Satoh T, Liao Y, Song C, Hu CD, Kariya KK, et al. Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras. J Biol Chem. 2001; 276:42219–42225. PMID: 11524421.
Article14. Iwasaki M, Tanaka R, Hishiya A, Homma S, Reed JC, Takayama S. BAG3 directly associates with guanine nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell adhesion. Biochem Biophys Res Commun. 2010; 400:413–418. PMID: 20800573.
Article15. Moiola C, De Luca P, Gardner K, Vazquez E, De Siervi A. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle. 2010; 9:3119–3126. PMID: 20714219.
Article16. Munteanu AI. Genetic alterations in nuclear receptor coactivators in breast cancer [dissertation]. [Los Angeles, USA]: University of Southern California;2010.17. Park UH, Kang MR, Kim EJ, Kwon YS, Hur W, Yoon SK, et al. ASXL2 promotes proliferation of breast cancer cells by linking ER alpha to histone methylation. Oncogene. 2016; 35:3742–3752. PMID: 26640146.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Classification of Triple-Negative Breast Cancer
- Analysis of H3K4me3-ChIP-Seq and RNA-Seq data to understand the putative role of miRNAs and their target genes in breast cancer cell lines
- The Construction of Regulatory Network for Insulin-Mediated Genes by Integrating Methods Based on Transcription Factor Binding Motifs and Gene Expression Variations
- Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
- Integrative Analysis of Microarray Data to Reveal Regulation Patterns in the Pathogenesis of Hepatocellular Carcinoma