Ann Lab Med.
2019 May;39(3):299-310. 10.3343/alm.2019.39.3.299.
Chromosomal Microarray Analysis as a First-Tier Clinical Diagnostic Test in Patients With Developmental Delay/Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies: A Prospective Multicenter Study in Korea
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea. microkim@catholic.ac.kr
- 2Catholic Genetic Laboratory Center, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 3Department of Rehabilitation Medicine, Daejeon St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Daejeon, Korea.
- 4Department of Rehabilitation Medicine, St. Vincent's Hospital, College of Medicine, The Catholic University of Korea, Suwon, Korea.
- 5Department of Rehabilitation Medicine, Incheon St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Incheon, Korea.
- 6Department of Pediatrics, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 7Department of Pediatrics, St. Vincent's Hospital, College of Medicine, The Catholic University of Korea, Suwon, Korea.
- 8Department of Pediatrics, Yeouido St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 9Department of Laboratory Medicine, Inha University School of Medicine, Incheon, Korea.
- 10Department of Rehabilitation Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 2431608
- DOI: http://doi.org/10.3343/alm.2019.39.3.299
Abstract
- BACKGROUND
To validate the clinical application of chromosomal microarray analysis (CMA) as a first-tier clinical diagnostic test and to determine the impact of CMA results on patient clinical management, we conducted a multicenter prospective study in Korean patients diagnosed as having developmental delay/intellectual disability (DD/ID), autism spectrum disorders (ASD), and multiple congenital anomalies (MCA).
METHODS
We performed both CMA and G-banding cytogenetics as the first-tier tests in 617 patients. To determine whether the CMA results directly influenced treatment recommendations, the referring clinicians were asked to complete a 39-item questionnaire for each patient separately after receiving the CMA results.
RESULTS
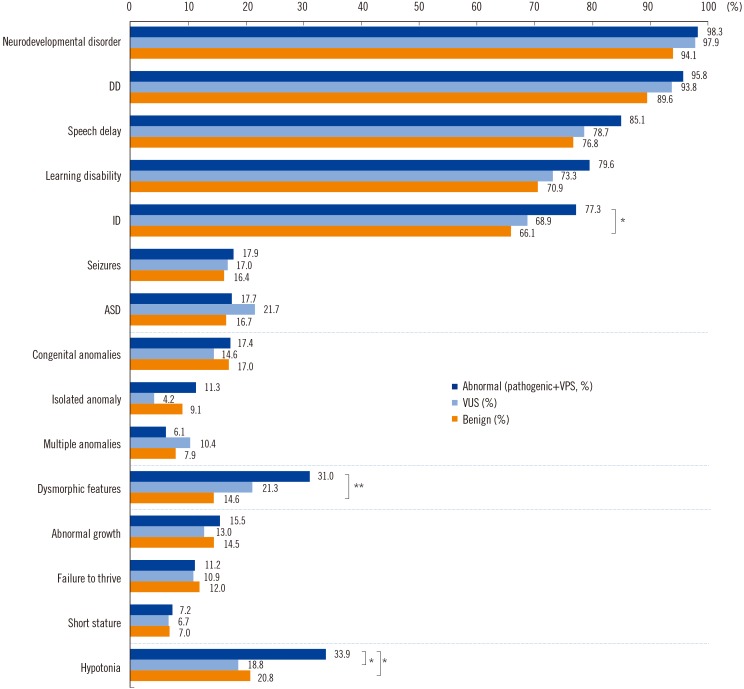
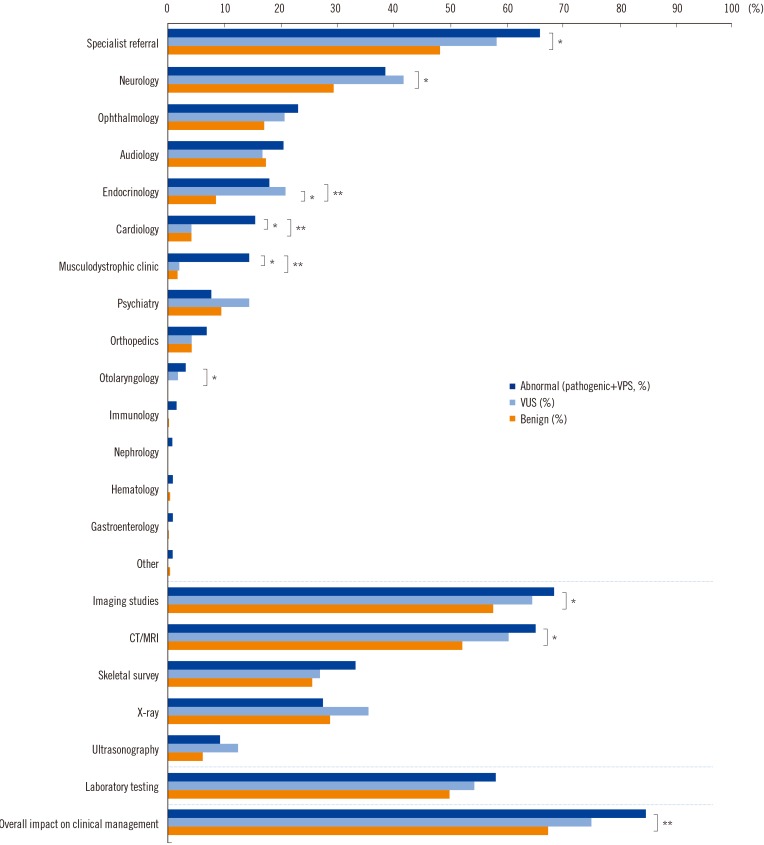
A total of 122 patients (19.8%) had abnormal CMA results, with either pathogenic variants (N=65) or variants of possible significance (VPS, N=57). Thirty-five well-known diseases were detected: 16p11.2 microdeletion syndrome was the most common, followed by Prader-Willi syndrome, 15q11-q13 duplication, Down syndrome, and Duchenne muscular dystrophy. Variants of unknown significance (VUS) were discovered in 51 patients (8.3%). VUS of genes putatively associated with developmental disorders were found in five patients: IMMP2L deletion, PTCH1 duplication, and ATRNL1 deletion. CMA results influenced clinical management, such as imaging studies, specialist referral, and laboratory testing in 71.4% of patients overall, and in 86.0%, 83.3%, 75.0%, and 67.3% of patients with VPS, pathogenic variants, VUS, and benign variants, respectively.
CONCLUSIONS
Clinical application of CMA as a first-tier test improves diagnostic yields and the quality of clinical management in patients with DD/ID, ASD, and MCA.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
Clinical Application of Sequential Epigenetic Analysis for Diagnosis of Silver–Russell Syndrome
Soo Yeon Kim, Chang Ho Shin, Young Ah Lee, Choong Ho Shin, Sei Won Yang, Tae-Joon Cho, Jung Min Ko
Ann Lab Med. 2021;41(4):401-408. doi: 10.3343/alm.2021.41.4.401.Clinical Utility of Methylation-Specific Multiplex Ligation-Dependent Probe Amplification for the Diagnosis of Prader–Willi Syndrome and Angelman Syndrome
Boram Kim, Yongsook Park, Sung Im Cho, Man Jin Kim, Jong-Hee Chae, Ji Yeon Kim, Moon-Woo Seong, Sung Sup Park
Ann Lab Med. 2022;42(1):79-88. doi: 10.3343/alm.2022.42.1.79.
Reference
-
1. Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011; 45:203–226. PMID: 21854229.2. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010; 86:749–764. PMID: 20466091.3. Manning M, Hudgins L. Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010; 12:742–745. PMID: 20962661.4. Sanmann JN, Pickering DL, Golden DM, Stevens JM, Hempel TE, Althof PA, et al. Assessing the utility of confirmatory studies following identification of large-scale genomic imbalances by microarray. Genet Med. 2015; 17:875–879. PMID: 25590977.5. Haddow JE, Palomaki GE. ACCE: a model process for evaluating data on emerging genetic tests. In : Khoury MJ, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. New York: Oxford University Press;2004. p. 217–233.6. Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DA. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med. 2014; 16:657–664. PMID: 24625444.7. Riggs ER, Wain KE, Riethmaier D, Smith-Packard B, Faucett WA, Hoppman N, et al. Chromosomal microarray impacts clinical management. Clin Genet. 2014; 85:147–153. PMID: 23347240.8. Coulter ME, Miller DT, Harris DJ, Hawley P, Picker J, Roberts AE, et al. Chromosomal microarray testing influences medical management. Genet Med. 2011; 13:770–776. PMID: 21716121.9. Tonelli M, Parkin P, Brauer P, Leduc D, Pottie K, Jaramillo Garcia A, et al. Recommendations on screening for developmental delay. CMAJ. 2016; 188:579–587. PMID: 27026672.10. Council on Children with Disabilities. Section on Developmental Behavioral Pediatrics. Bright Futures Steering Committee. Medical Home Initiatives for Children With Special Needs Project Advisory Committee. Identifying infants and young children with developmental disorders in the medical home: an algorithm for developmental surveillance and screening. Pediatrics. 2006; 118:405–420. PMID: 16818591.11. McGowan-Jordan J, Simons A, Schmid M, editors. ISCN : an international system for human cytogenomic nomenclature (2016). Basel: Karger;2016.12. Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011; 13:680–685. PMID: 21681106.13. Bartnik M, Wiśniowiecka-Kowalnik B, Nowakowska B, Smyk M, Kędzior M, Sobecka K, et al. The usefulness of array comparative genomic hybridization in clinical diagnostics of intellectual disability in children. Dev Period Med. 2014; 18:307–317. PMID: 25182394.14. Cappuccio G, Vitiello F, Casertano A, Fontana P, Genesio R, Bruzzese D, et al. New insights in the interpretation of array-CGH: autism spectrum disorder and positive family history for intellectual disability predict the detection of pathogenic variants. Ital J Pediatr. 2016; 42:39. PMID: 27072107.15. Ozyilmaz B, Kirbiyik O, Koc A, Ozdemir TR, Kaya OO, Guvenc MS, et al. Experiences in microarray-based evaluation of developmental disabilities and congenital anomalies. Clin Genet. 2017; 92:372–379. PMID: 28128450.16. Xiang B, Zhu H, Shen Y, Miller DT, Lu K, Hu X, et al. Genome-wide oligonucleotide array comparative genomic hybridization for etiological diagnosis of mental retardation: a multicenter experience of 1499 clinical cases. J Mol Diagn. 2010; 12:204–212. PMID: 20093387.17. Shin S, Yu N, Choi JR, Jeong S, Lee KA. Routine chromosomal microarray analysis is necessary in Korean patients with unexplained developmental delay/mental retardation/autism spectrum disorder. Ann Lab Med. 2015; 35:510–518. PMID: 26206688.18. Glancy M, Barnicoat A, Vijeratnam R, de Souza S, Gilmore J, Huang S, et al. Transmitted duplication of 8p23.1-8p23.2 associated with speech delay, autism and learning difficulties. Eur J Hum Genet. 2009; 17:37–43. PMID: 18716609.19. Harada N, Takano J, Kondoh T, Ohashi H, Hasegawa T, Sugawara H, et al. Duplication of 8p23.2: a benign cytogenetic variant? Am J Med Genet. 2002; 111:285–288. PMID: 12210324.20. Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen–the Clinical Genome Resource. N Engl J Med. 2015; 372:2235–2242. PMID: 26014595.21. Bertelsen B, Melchior L, Jensen LR, Groth C, Glenthøj B, Rizzo R, et al. Intragenic deletions affecting two alternative transcripts of the IMMP2L gene in patients with Tourette syndrome. Eur J Hum Genet. 2014; 22:1283–1289. PMID: 24549057.22. Gimelli S, Capra V, Di Rocco M, Leoni M, Mirabelli-Badenier M, Schiaffino MC, et al. Interstitial 7q31.1 copy number variations disrupting IMMP2L gene are associated with a wide spectrum of neurodevelopmental disorders. Mol Cytogenet. 2014; 7:54. PMID: 25478008.23. Derwińska K, Smyk M, Cooper ML, Bader P, Cheung SW, Stankiewicz P. PTCH1 duplication in a family with microcephaly and mild developmental delay. Eur J Hum Genet. 2009; 17:267–271. PMID: 18830227.24. Stark Z, Bruno DL, Mountford H, Lockhart PJ, Amor DJ. De novo 325 kb microdeletion in chromosome band 10q25.3 including ATRNL1 in a boy with cognitive impairment, autism and dysmorphic features. Eur J Med Genet. 2010; 53:337–339. PMID: 20670697.25. Shoukier M, Klein N, Auber B, Wickert J, Schröder J, Zoll B, et al. Array CGH in patients with developmental delay or intellectual disability: are there phenotypic clues to pathogenic copy number variants? Clin Genet. 2013; 83:53–65. PMID: 22283495.26. Lee CG, Park SJ, Yun JN, Ko JM, Kim HJ, Yim SY, et al. Array-based comparative genomic hybridization in 190 Korean patients with developmental delay and/or intellectual disability: a single tertiary care university center study. Yonsei Med J. 2013; 54:1463–1470. PMID: 24142652.27. Hayeems RZ, Hoang N, Chenier S, Stavropoulos DJ, Pu S, Weksberg R, et al. Capturing the clinical utility of genomic testing: medical recommendations following pediatric microarray. Eur J Hum Genet. 2015; 23:1135–1141. PMID: 25491637.28. Saam J, Gudgeon J, Aston E, Brothman AR. How physicians use array comparative genomic hybridization results to guide patient management in children with developmental delay. Genet Med. 2008; 10:181–186. PMID: 18344707.29. Nevado J, Mergener R, Palomares-Bralo M, Souza KR, Vallespín E, Mena R, et al. New microdeletion and microduplication syndromes: a comprehensive review. Genet Mol Biol. 2014; 37:210–219. PMID: 24764755.30. Weise A, Mrasek K, Klein E, Mulatinho M, Llerena JC Jr, Hardekopf D, et al. Microdeletion and microduplication syndromes. J Histochem Cytochem. 2012; 60:346–358. PMID: 22396478.31. Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, Weiss ME, Köster J, Marais A, et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet. 2017; 25:176–182. PMID: 27848944.32. Grozeva D, Carss K, Spasic-Boskovic O, Tejada MI, Gecz J, Shaw M, et al. Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutat. 2015; 36:1197–1204. PMID: 26350204.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Chromosomal Microarray Testing in 42 Korean Patients with Unexplained Developmental Delay, Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies
- Diagnostic distal 16p11.2 deletion in a preterm infant with facial dysmorphism
- Chromosomal Microarray With Clinical Diagnostic Utility in Children With Developmental Delay or Intellectual Disability
- Clinical Applications of Chromosomal Microarray Analysis
- Routine Chromosomal Microarray Analysis is Necessary in Korean Patients With Unexplained Developmental Delay/Mental Retardation/Autism Spectrum Disorder