Chromosomal Microarray With Clinical Diagnostic Utility in Children With Developmental Delay or Intellectual Disability
- Affiliations
-
- 1Department of Pediatrics, Department of Genome Medicine and Science, Gil Medical Center, Gachon University College of Medicine, Incheon, Korea.
- 2Department of Pediatrics, Seoul National University Bundang Hospital, Seongnam, Korea.
- 3Department of Pediatrics, Pediatric Clinical Neuroscience Center, Seoul National University Children's Hospital, Seoul National University College of Medicine, Seoul, Korea. prabbit7@snu.ac.kr
- 4The Institute of Reproductive Medicine and Population, Medical Research Center, Seoul National University College of Medicine, Seoul, Korea.
- 5Department of Obstetrics and Gynecology, Seoul National University Hospital, Seoul, Korea.
- KMID: 2434738
- DOI: http://doi.org/10.3343/alm.2018.38.5.473
Abstract
- BACKGROUND
Chromosomal microarray (CMA) testing is a first-tier test for patients with developmental delay, autism, or congenital anomalies. It increases diagnostic yield for patients with developmental delay or intellectual disability. In some countries, including Korea, CMA testing is not yet implemented in clinical practice. We assessed the diagnostic utility of CMA testing in a large cohort of patients with developmental delay or intellectual disability in Korea.
METHODS
We conducted a genome-wide microarray analysis of 649 consecutive patients with developmental delay or intellectual disability at the Seoul National University Children's Hospital. Medical records were reviewed retrospectively. Pathogenicity of detected copy number variations (CNVs) was evaluated by referencing previous reports or parental testing using FISH or quantitative PCR.
RESULTS
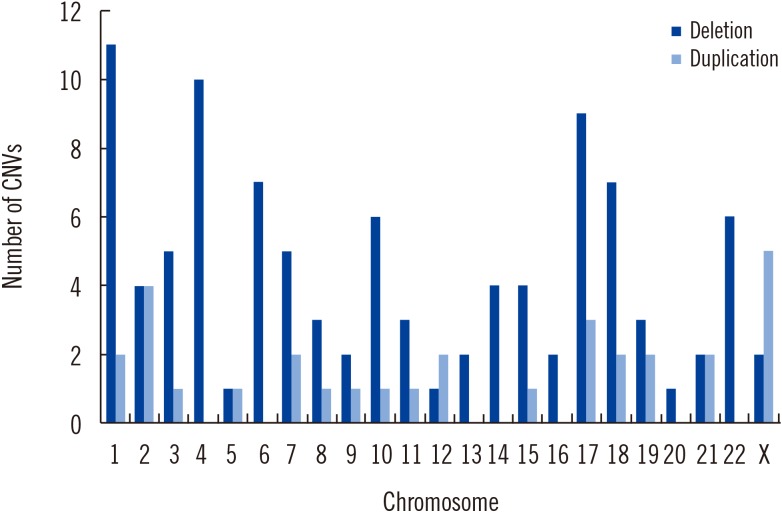
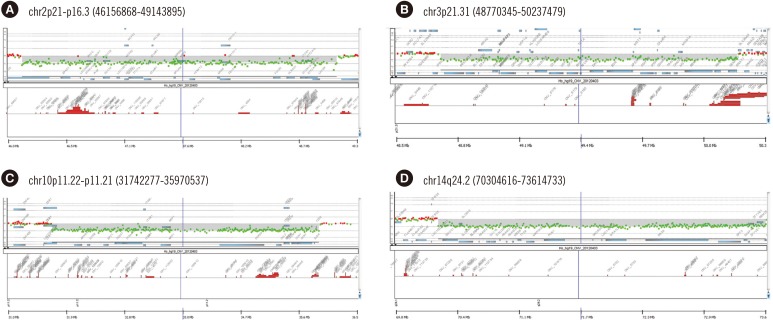
We found 110 patients to have pathogenic CNVs, which included 100 deletions and 31 duplications of 270 kb to 30 Mb. The diagnostic yield was 16.9%, demonstrating the diagnostic utility of CMA testing in clinic. Parental testing was performed in 66 patients, 86.4% of which carried de novo CNVs. In eight patients, pathogenic CNVs were inherited from healthy parents with a balanced translocation, and genetic counseling was provided to these families. We verified five rarely reported deletions on 2p21p16.3, 3p21.31, 10p11.22, 14q24.2, and 21q22.13.
CONCLUSIONS
This study demonstrated the clinical utility of CMA testing in the genetic diagnosis of patients with developmental delay or intellectual disability. CMA testing should be included as a clinical diagnostic test for all children with developmental delay or intellectual disability.
Keyword
MeSH Terms
Figure
Cited by 3 articles
-
Low-Depth 전장유전체염기서열검사 기반 유전자 복제수변이 분석을 이용한
MECP2 duplication의 진단
Mi-Ae Jang, Soyoung Park, Jong Eun Park, Young-Eun Kim, Chang-Seok Ki
Lab Med Online. 2020;10(2):165-168. doi: 10.3343/lmo.2020.10.2.165.Clinical Application of Sequential Epigenetic Analysis for Diagnosis of Silver–Russell Syndrome
Soo Yeon Kim, Chang Ho Shin, Young Ah Lee, Choong Ho Shin, Sei Won Yang, Tae-Joon Cho, Jung Min Ko
Ann Lab Med. 2021;41(4):401-408. doi: 10.3343/alm.2021.41.4.401.Kleefstra syndrome combined with vesicoureteral reflux and rectourethral fistula: a case report and literature review
Chae Won Lee, Min Ji Park, Eun Joo Lee, Sangyoon Lee, Jinyoung Park, Jun Nyung Lee, So Mi Lee, Shin Young Jeong, Min Hyun Cho
Child Kidney Dis. 2022;26(2):111-115. doi: 10.3339/ckd.22.039.
Reference
-
1. Shevell M, Ashwal S, Donley D, Flint J, Gingold M, Hirtz D, et al. Practice parameter: evaluation of the child with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and The Practice Committee of the Child Neurology Society. Neurology. 2003; 60:367–380. PMID: 12578916.2. Riggs ER, Wain KE, Riethmaier D, Smith-Packard B, Faucett WA, Hoppman N, et al. Chromosomal microarray impacts clinical management. Clin Genet. 2014; 85:147–153. PMID: 23347240.3. Tao VQ, Chan KY, Chu YW, Mok GT, Tan TY, Yang W, et al. The clinical impact of chromosomal microarray on paediatric care in Hong Kong. PLoS One. 2014; 9:e109629. PMID: 25333781.4. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010; 86:749–764. PMID: 20466091.5. Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011; 13:680–685. PMID: 21681106.6. Trakadis Y, Shevell M. Microarray as a first genetic test in global developmental delay: a cost-effectiveness analysis. Dev Med Child Neurol. 2011; 53:994–999. PMID: 21848878.7. Shin S, Yu N, Choi JR, Jeong S, Lee KA. Routine chromosomal microarray analysis is necessary in Korean patients with unexplained developmental delay/mental retardation/autism spectrum disorder. Ann Lab Med. 2015; 35:510–518. PMID: 26206688.8. Sharma P, Gupta N, Chowdhury MR, Sapra S, Ghosh M, Gulati S, et al. Application of chromosomal microarrays in the evaluation of intellectual disability/global developmental delay patients-A study from a tertiary care genetic centre in India. Gene. 2016; 590:109–119. PMID: 27291820.9. Lee CG, Park SJ, Yun JN, Ko JM, Kim HJ, Yim SY, et al. Array-based comparative genomic hybridization in 190 Korean patients with developmental delay and/or intellectual disability: a single tertiary care university center study. Yonsei Med J. 2013; 54:1463–1470. PMID: 24142652.10. Byeon JH, Shin E, Kim GH, Lee K, Hong YS, Lee JW, et al. Application of array-based comparative genomic hybridization to pediatric neurologic diseases. Yonsei Med J. 2014; 55:30–36. PMID: 24339284.11. ISCN. Shaffer LG, McGowan-Jordan J, Schmid M, editors. An international system for human cytogenetic nomenclature. Basel: S. Karger;2013.12. ISCN. McGowan-Jordan J, Simons A, Schmid M, editors. An international system for human cytogenetic nomenclature. Basel: S. Karger;2016.13. Di Gregorio E, Savin E, Biamino E, Belligni EF, Naretto VG, d'Alessandro G, et al. Large cryptic genomic rearrangements with apparently normal karyotypes detected by array-CGH. Mol Cytogenet. 2014; 7:82. PMID: 25435912.14. Siggberg L, Ala-Mello S, Jaakkola E, Kuusinen E, Schuit R, Kohlhase J, et al. Array CGH in molecular diagnosis of mental retardation-a study of 150 Finnish patients. Am J Med Genet A. 2010; 152a:1398–1410. PMID: 20503314.15. Chong WW, Lo IF, Lam ST, Wang CC, Luk HM, Leung TY, et al. Performance of chromosomal microarray for patients with intellectual disabilities/developmental delay, autism, and multiple congenital anomalies in a Chinese cohort. Mol Cytogenet. 2014; 7:34. PMID: 24926319.16. Xiang B, Zhu H, Shen Y, Miller DT, Lu K, Hu X, et al. Genome-wide oligonucleotide array comparative genomic hybridization for etiological diagnosis of mental retardation: a multicenter experience of 1,499 clinical cases. J Mol Diagn. 2010; 12:204–212. PMID: 20093387.17. Fan YS, Jayakar P, Zhu H, Barbouth D, Sacharow S, Morales A, et al. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007; 28:1124–1132. PMID: 17621639.18. Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, Capalbo A, et al. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur J Paediatr Neurol. 2013; 17:589–599. PMID: 23711909.19. Pickering DL, Eudy JD, Olney AH, Dave BJ, Golden D, Stevens J, et al. Array-based comparative genomic hybridization analysis of 1,176 consecutive clinical genetics investigations. Genet Med. 2008; 10:262–266. PMID: 18414209.20. D'Arrigo S, Gavazzi F, Alfei E, Zuffardi O, Montomoli C, Corso B, et al. The diagnostic yield of array comparative genomic hybridization is high regardless of severity of intellectual disability/developmental delay in children. J Child Neurol. 2016; 31:691–699. PMID: 26511719.21. Shoukier M, Klein N, Auber B, Wickert J, Schröder J, Zoll B, et al. Array CGH in patients with developmental delay or intellectual disability: are there phenotypic clues to pathogenic copy number variants? Clin Genet. 2013; 83:53–65. PMID: 22283495.22. Shimada S, Shimojima K, Okamoto N, Sangu N, Hirasawa K, Matsuo M, et al. Microarray analysis of 50 patients reveals the critical chromosomal regions responsible for 1p36 deletion syndrome-related complications. Brain Dev. 2015; 37:515–526. PMID: 25172301.23. Battaglia A. 1p36 Deletion Syndrome. In : Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, editors. GeneReviews. Seattle: University of Washington;2008.24. Battaglia A, Carey JC, South ST. Wolf-Hirschhorn Syndrome. In : Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, editors. GeneReviews. Seattle: University of Washington;2002.25. Bertini V, De Vito G, Costa R, Simi P, Valetto A. Isolated 6q terminal deletions: an emerging new syndrome. Am J Med Genet A. 2006; 140:74–81. PMID: 16329114.26. Lin S, Zhou Y, Fang Q, Wu J, Zhang Z, Ji Y, et al. Chromosome 10q26 deletion syndrome: two new cases and a review of the literature. Mol Med Rep. 2016; 14:5134–5140. PMID: 27779662.27. Smith ACM, Boyd KE, et al. Smith-Magenis Syndrome. In : Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, editors. GeneReviews. Seattle: University of Washington;2001.28. Maya I, Vinkler C, Konen O, Kornreich L, Steinberg T, Yeshaya J, et al. Abnormal brain magnetic resonance imaging in two patients with Smith-Magenis syndrome. Am J Med Genet A. 2014; 164a:1940–1946. PMID: 24788350.29. McDonald-McGinn DM, Emanuel BS, Zackai EH. 22q11.2 Deletion syndrome. In : Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mefford HC, editors. GeneReviews. Seattle: University of Washington;1999.30. Bohm LA, Zhou TC, Mingo TJ, Dugan SL, Patterson RJ, Sidman JD, et al. Neuroradiographic findings in 22q11.2 deletion syndrome. Am J Med Genet A. 2017; 173:2158–2165. PMID: 28577347.31. Briggs TA, Harris J, Innes J, Will A, Arkwright PD, Clayton-Smith J. The value of microarray-based comparative genomic hybridisation (aCGH) testing in the paediatric clinic. Arch Dis Child. 2015; 100:728–731. PMID: 25809346.32. Roselló M, Martínez F, Monfort S, Mayo S, Oltra S, Orellana C. Phenotype profiling of patients with intellectual disability and copy number variations. Eur J Paediatr Neurol. 2014; 18:558–566. PMID: 24815074.33. Buizer-Voskamp JE, Blauw HM, Boks MP, van Eijk KR, Veldink JH, Hennekam EA, et al. Increased paternal age and the influence on burden of genomic copy number variation in the general population. Hum Genet. 2013; 132:443–450. PMID: 23315237.34. Nowakowska BA, de Leeuw N, Ruivenkamp CA, Sikkema-Raddatz B, Crolla JA, Thoelen R, et al. Parental insertional balanced translocations are an important cause of apparently de novo CNVs in patients with developmental anomalies. Eur J Hum Genet. 2012; 20:166–170. PMID: 21915152.35. Sanders SR, Dawson AJ, Vust A, Hryshko M, Tomiuk M, Riordan D, et al. Interstitial deletion of chromosome 2p16.2p21. Clin Dysmorphol. 2003; 12:183–185. PMID: 14564157.36. Eto K, Sakai N, Shimada S, Shioda M, Ishigaki K, Hamada Y, et al. Microdeletions of 3p21.31 characterized by developmental delay, distinctive features, elevated serum creatine kinase levels, and white matter involvement. Am J Med Genet A. 2013; 161A:3049–3056. PMID: 24039031.37. Haldeman-Englert CR, Gai X, Perin JC, Ciano M, Halbach SS, Geiger EA, et al. A 3.1-Mb microdeletion of 3p21.31 associated with cortical blindness, cleft lip, CNS abnormalities, and developmental delay. Eur J Med Genet. 2009; 52:265–268. PMID: 19100872.38. Tassano E, Accogli A, Panigada S, Ronchetto P, Cuoco C, Gimelli G. Phenotypic and genetic characterization of a patient with a de novo interstitial 14q24.1q24.3 deletion. Mol Cytogenet. 2014; 7:49. PMID: 25076984.39. Ji J, Lee H, Argiropoulos B, Dorrani N, Mann J, Martinez-Agosto JA, et al. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur J Hum Genet. 2015; 23:1473–1481. PMID: 25944381.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Chromosomal Microarray Testing in 42 Korean Patients with Unexplained Developmental Delay, Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies
- Phenotypic Analysis of Korean Patients with Abnormal Chromosomal Microarray in Patients with Unexplained Developmental Delay/Intellectual Disability
- The First Cases of OPHN1 Exons 1 and 2 Deletion in Two X-linked Intellectual Developmental Disorder Patients in Korea
- A Cytogenetic Study in 120 Turkish Children with Intellectual Disability and Characteristics of Fragile X Syndrome
- Diagnostic distal 16p11.2 deletion in a preterm infant with facial dysmorphism