Ann Lab Med.
2018 Nov;38(6):563-568. 10.3343/alm.2018.38.6.563.
Delamanid, Bedaquiline, and Linezolid Minimum Inhibitory Concentration Distributions and Resistance-related Gene Mutations in Multidrug-resistant and Extensively Drug-resistant Tuberculosis in Korea
- Affiliations
-
- 1Department of R&D, Korean Institute of Tuberculosis, Cheongju, Korea. seung6992@hanmail.net
- KMID: 2429122
- DOI: http://doi.org/10.3343/alm.2018.38.6.563
Abstract
- BACKGROUND
Delamanid, bedaquiline, and linezolid have recently been approved for the treatment of multidrug- and extensively drug-resistant (MDR and XDR, respectively) tuberculosis (TB). To use these drugs effectively, drug susceptibility tests, including rapid molecular techniques, are required for accurate diagnosis and treatment. Furthermore, mutation analyses are needed to assess the potential for resistance. We evaluated the minimum inhibitory concentrations (MICs) of these three anti-TB drugs for Korean MDR and XDR clinical strains and mutations in genes related to resistance to these drugs.
METHODS
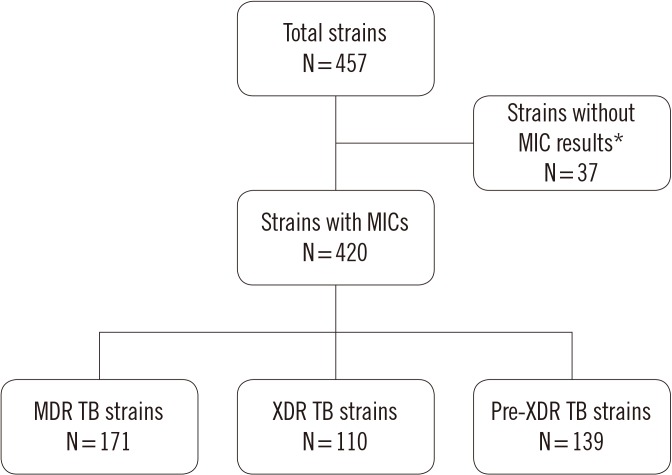
MICs were determined for delamanid, bedaquiline, and linezolid using a microdilution method. The PCR products of drug resistance-related genes from 420 clinical Mycobacterium tuberculosis strains were sequenced and aligned to those of M. tuberculosis H37Rv.
RESULTS
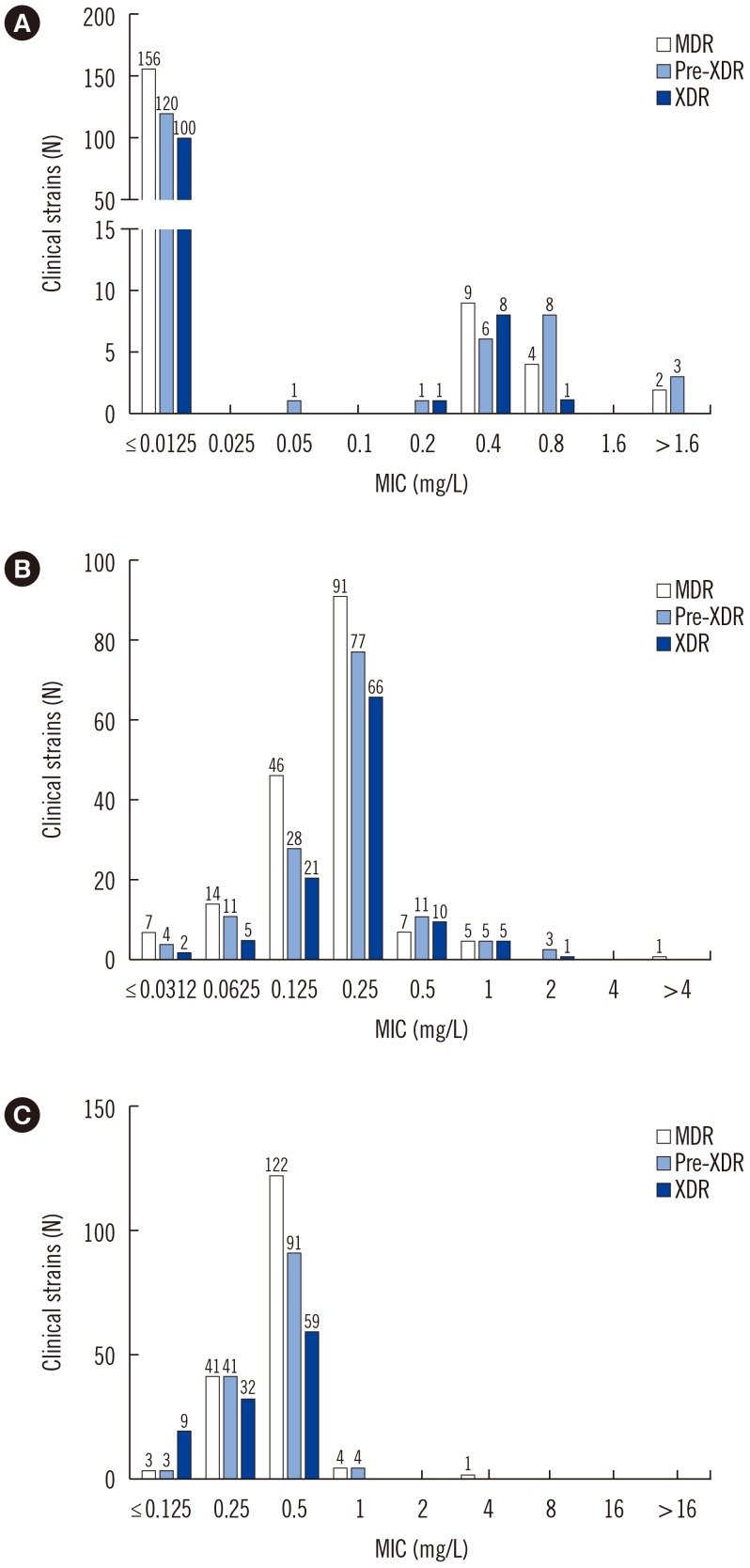
The overall MICs for delamanid, bedaquiline, and linezolid ranged from ≤0.025 to >1.6 mg/L, ≤0.0312 to >4 mg/L, and ≤0.125 to 1 mg/L, respectively. Numerous mutations were found in drug-susceptible and -resistant strains. We did not detect specific mutations associated with resistance to bedaquiline and linezolid. However, the Gly81Ser and Gly81Asp mutations were associated with resistance to delamanid.
CONCLUSIONS
We determined the MICs of three anti-TB drugs for Korean MDR and XDR strains and identified various mutations in resistance-related genes. Further studies are needed to determine the genetic mechanisms underlying resistance to these drugs.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Additional Drug Resistance in Patients with Multidrug-resistant Tuberculosis in Korea: a Multicenter Study from 2010 to 2019
Taehoon Lee, Seung Jun Lee, Doosoo Jeon, Ho Young Lee, Hyo-Jung Kim, Bo Hyoung Kang, Jeongha Mok
J Korean Med Sci. 2021;36(26):e174. doi: 10.3346/jkms.2021.36.e174.
Reference
-
1. Blair HA, Scott LJ. Delamanid: a review of its use in patients with multidrug-resistant tuberculosis. Drugs. 2015; 75:91–100. PMID: 25404020.2. World Health Organization. Global tuberculosis report 2016. Updated on Jun 2016. http://www.who.int/tb/publications/global_report/en.3. Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, et al. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med. 2012; 366:2151–2160. PMID: 22670901.4. Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006; 3:e466. PMID: 17132069.5. Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, et al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015; 59:5316–5323. PMID: 26100695.6. Feuerriegel S, Köser CU, Baù D, Rüsch-Gerdes S, Summers DK, Archer JA, et al. Impact of fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother. 2011; 55:5718–5722. PMID: 21930879.7. Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005; 307:223–227. PMID: 15591164.8. Huitric E, Verhasselt P, Andries K, Hoffner SE. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2007; 51:4202–4204. PMID: 17709466.9. Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One. 2014; 9:e102135. PMID: 25010492.10. Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, Andersson DI. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2010; 54:1022–1028. PMID: 20038615.11. Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrob Chemother. 2013; 68:4–11. PMID: 22949625.12. Birmingham MC, Rayner CR, Meagher AK, Flavin SM, Batts DH, Schentag JJ. Linezolid for the treatment of multidrug-resistant, gram-positive infections: experience from a compassionate-use program. Clin Infect Dis. 2003; 36:159–168. PMID: 12522747.13. Hillemann D, Rüsch-Gerdes S, Richter E. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2008; 52:800–801. PMID: 18070973.14. Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, et al. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother. 2012; 56:2743–2745. PMID: 22371899.15. Schena E, Nedialkova L, Borroni E, Battaglia S, Cabibbe AM, Niemann S, et al. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtiter assay and the BACTEC™ MGIT™ 960 system. J Antimicrob Chemother. 2016; 71:1532–1539. PMID: 27076101.16. Kaniga K, Cirillo DM, Hoffner S, Ismail NA, Kaur D, Lounis N, et al. A multilaboratory, multicountry study to determine bedaquiline minimal inhibitory concentration quality control ranges for phenotypic drug-susceptibility testing. J Clin Microbiol. 2016; 54:2956–2962. PMID: 27654337.17. Keller PM, Hömke R, Ritter C, Valsesia G, Bloemberg GV, Bötteger EC. Determination of MIC distribution and epidemiological cutoff values for bedaquiline and delamanid in Mycobacterium tuberculosis using the MGIT 960 system equipped with TB eXiST. Antimicrob Agents Chemother. 2015; 59:4352–4355. PMID: 25941226.18. Bloemberg GV, Keller PM, Stucki D, Trauner A, Borrell S, Latshang T, et al. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med. 2015; 373:1986–1988. PMID: 26559594.19. Huang TS, Liu YC, Sy CL, Chen YS, Tu HZ, Chen BC. In vitro activities of linezolid against clinical isolates of Mycobacterium tuberculosis complex isolated in Taiwan over 10 years. Antimicrob Agents Chemother. 2008; 52:2226–2227. PMID: 18391030.20. Lee SH, Choi HB, Yu SY, Chang UJ, Kim CK, Kim HJ. Detection of first-line anti-tuberculosis drug resistance mutations by allele-specific primer extension on a microsphere-based platform. Ann Lab Med. 2015; 35:487–493. PMID: 26206684.21. Stinson K, Kurepina N, Venter A, Fujiwara M, Kawasaki M, Timm J, et al. MIC of delamanid (OPC-67683) against Mycobacterium tuberculosis clinical isolates and a proposed critical concentration. Antimicrob Agents Chemother. 2016; 60:3316–3322. PMID: 26976868.22. European Committee on Antimicrobial Susceptibility Testing. Definitions of clinical breakpoints and epidemiological cut-off values. Clinical Breakpoints Table V.7.1. Valid From 2017-03-10. Updated on Mar 2017. http://www.eucast.org/clinical_breakpoints/.23. CLSI. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. 2nd edition. Wayne, PA: Clinical Laboratory Standards Institute;2011. CLSI M24-A2.24. Segala E, Sougakoff W, Chauffour AN, Jarller V, Petrella S. New mutations in the mycobacterial ATP synthase: new insights into the binding of the diarylquinoline TMC207 to the ATP synthase C-ring structure. Antimicrob Agents Chemother. 2012; 56:2326–2334. PMID: 22354303.25. Zhang S, Chen J, Cui P, Shi W, Shi X, Niu H, et al. Mycobacterium tuberculosis mutations associated with reduced susceptibility to linezolid. Antimicrob Agents Chemother. 2016; 60:2542–2544. PMID: 26810645.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Erratum: Delamanid, Bedaquiline, and Linezolid Minimum Inhibitory Concentration Distributions and Resistance-related Gene Mutations in Multidrug-resistant and Extensively Drug-resistant Tuberculosis in Korea
- Concurrent, Prolonged Use of Bedaquiline and Delamanid for Multidrug-Resistant Tuberculosis
- Shorter Oral Regimen for Multidrug Resistant Tuberculosis in South Korea
- Medical Treatment of Pulmonary Multidrug-Resistant Tuberculosis
- Concise Clinical Review of Hematologic Toxicity of Linezolid in Multidrug-Resistant and Extensively Drug-Resistant Tuberculosis: Role of Mitochondria