J Vet Sci.
2018 Sep;19(5):627-634. 10.4142/jvs.2018.19.5.627.
Genetic diversity of bovine Mycobacterium avium subsp. paratuberculosis discriminated by IS1311 PCR-REA, MIRU-VNTR, and MLSSR genotyping
- Affiliations
-
- 1Department of Infectious Disease, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea. yoohs@snu.ac.kr
- 2Department of Animal Resources Development, National Institute of Animal Science, Rural Development Administration, Cheonan 31000, Korea.
- 3Institute of Green Bio Science and Technology, Seoul National University, Pyeongchang 25354, Korea.
- KMID: 2420931
- DOI: http://doi.org/10.4142/jvs.2018.19.5.627
Abstract
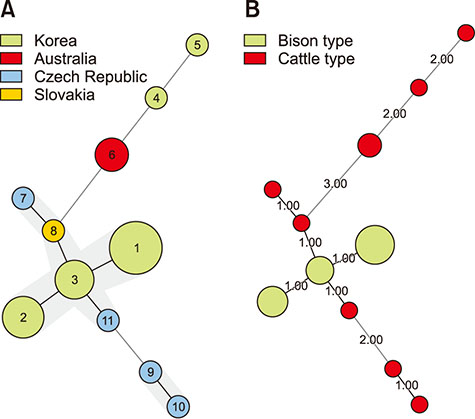
- The aim of this study was to describe the genetic diversity of Mycobacterium avium subsp. paratuberculosis (MAP) obtained from individual cows in Korea. Twelve MAP-positive fecal DNA samples and 19 MAP isolates were obtained from 10 cattle herds located in 5 provinces in Korea. In addition, 5 MAP isolates obtained from the Czech Republic and Slovakia and 3 isolates from Australia were genotyped for comparison with the domestic isolates. The most prevalent strains in Korea were of the "bison-type" genotype (23 of 31 fecal DNA/isolates) and were distributed nationwide. The remaining MAP isolates (8) and all of the foreign isolates were identified as "cattle-type". The bison-type strains which were discriminated only as INMV 68 in variable-number tandem repeats of mycobacterial interspersed repetitive units (MIRU-VNTR) typing. Multilocus short sequence repeat (MLSSR) typing differentiated the bison-type strains into 3 different subtypes. The cattle-type strains were divided into 3 subtypes by MIRU-VNTR and 8 subtypes by MLSSR. The allelic diversities in the MIRU-VNTR and MLSSR results were calculated as 0.567 and 0.866, respectively. These results suggest that MIRU-VNTR typing cannot provide a sufficient description of the epidemiological situation of MAP. Therefore, an alternative method, such as MLSSR, is needed for typing of MAP strains to elucidate the molecular epidemiology of MAP infections. Overall, this study is the first epidemiological survey report in Korea using both MIRU-VNTR and MLSSR typing methods, and it has provided basic data necessary to elucidate the characteristics of MAP infections in Korea.
MeSH Terms
Figure
Reference
-
1. Ahlstrom C, Barkema HW, Stevenson K, Zadoks RN, Biek R, Kao R, Trewby H, Haupstein D, Kelton DF, Fecteau G, Labrecque O, Keefe GP, McKenna SL, De Buck J. Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level. BMC Genomics. 2015; 16:161.2. Amonsin A, Li LL, Zhang Q, Bannantine JP, Motiwala AS, Sreevatsan S, Kapur V. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J Clin Microbiol. 2004; 42:1694–1702.
Article3. Bryant JM, Thibault VC, Smith DG, McLuckie J, Heron I, Sevilla IA, Biet F, Harris SR, Maskell DJ, Bentley SD, Parkhill J, Stevenson K. Phylogenomic exploration of the relationships between strains of Mycobacterium avium subspecies paratuberculosis. BMC Genomics. 2016; 17:79.
Article4. Corbett CS, De Buck J, Orsel K, Barkema HW. Fecal shedding and tissue infections demonstrate transmission of Mycobacterium avium subsp. paratuberculosis in group-housed dairy calves. Vet Res. 2017; 48:27.
Article5. Coussens PM. Mycobacterium paratuberculosis and the bovine immune system. Anim Health Res Rev. 2001; 2:141–161.6. de Kruijf M, Lesniak ON, Yearsley D, Ramovic E, Coffey A, O'Mahony J. Low genetic diversity of bovine Mycobacterium avium subspecies paratuberculosis isolates detected by MIRU-VNTR genotyping. Vet Microbiol. 2017; 203:280–285.
Article7. Douarre PE, Cashman W, Buckley J, Coffey A, O'Mahony J. Molecular characterization of Mycobacterium avium subsp. paratuberculosis using multi-locus short sequence repeat (MLSSR) and mycobacterial interspersed repetitive units-variable number tandem repeat (MIRU-VNTR) typing methods. Vet Microbiol. 2011; 149:482–487.
Article8. Fernández-Silva JA, Abdulmawjood A, Akineden Ö, Dräger K, Klawonn W, Bülte M. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis at a regional scale in Germany. Res Vet Sci. 2012; 93:776–782.
Article9. Gioffré A, Correa Muñoz M, Alvarado Pinedo MF, Vaca R, Morsella C, Fiorentino MA, Paolicchi F, Ruybal P, Zumárraga M, Travería GE, Romano MI. Molecular typing of Argentinian Mycobacterium avium subsp. paratuberculosis isolates by multiple-locus variable number-tandem repeat analysis. Braz J Microbiol. 2015; 46:557–564.
Article10. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988; 26:2465–2466.
Article11. Imperiale BR, Moyano RD, DI Giulio AB, Romero MA, Alvarado Pinedo MF, Santangelo MP, Travería GE, Morcillo NS, Romano MI. Genetic diversity of Mycobacterium avium complex strains isolated in Argentina by MIRU-VNTR. Epidemiol Infect. 2017; 145:1382–1391.
Article12. Kasnitz N, Köhler H, Weigoldt M, Gerlach GF, Möbius P. Stability of genotyping target sequences of Mycobacterium avium subsp. paratuberculosis upon cultivation on different media, in vitro- and in vivo passage, and natural infection. Vet Microbiol. 2013; 167:573–583.
Article13. Kim JM, Ku BK, Lee HN, Hwang IY, Jang YB, Kim J, Hyun BH, Jung SC. Mycobacterium avium paratuberculosis in wild boars in Korea. J Wildl Dis. 2013; 49:413–417.14. Leão C, Goldstone RJ, Bryant J, McLuckie J, Inácio J, Smith DG, Stevenson K. Novel single nucleotide polymorphism-based assay for genotyping Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol. 2016; 54:556–564.
Article15. Marsh I, Whittington R, Cousins D. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol Cell Probes. 1999; 13:115–126.16. Möbius P, Luyven G, Hotzel H, Köhler H. High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J Clin Microbiol. 2008; 46:972–981.
Article17. Mortier RA, Barkema HW, Bystrom JM, Illanes O, Orsel K, Wolf R, Atkins G, De Buck J. Evaluation of age-dependent susceptibility in calves infected with two doses of Mycobacterium avium subspecies paratuberculosis using pathology and tissue culture. Vet Res. 2013; 44:94.18. Nielsen SS, Toft N. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev Vet Med. 2009; 88:1–14.
Article19. Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med. 1999; 40:179–192.
Article20. Park HT, Shin MK, Park HE, Cho YI, Yoo HS. PCR-based detection of Mycobacterium avium subsp. paratuberculosis infection in cattle in South Korea using fecal samples. J Vet Med Sci. 2016; 78:1537–1540.
Article21. Park HT, Shin MK, Sung KY, Park HE, Cho YI, Yoo HS. Effective DNA extraction method to improve detection of Mycobacterium avium subsp. paratuberculosis in bovine feces. Korean J Vet Res. 2014; 54:55–57.
Article22. Sevilla Ix, Singh SV, Garrido JM, Aduriz G, Rodríguez S, Geijo MV, Whittington RJ, Saunders V, Whitlock RH, Juste RA. Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions. Rev Sci Tech. 2005; 24:1061–1066.
Article23. Singh AV, Singh SV, Singh PK, Sohal JS. Genotype diversity in Indian isolates of Mycobacterium avium subspecies paratuberculosis recovered from domestic and wild ruminants from different agro-climatic regions. Comp Immunol Microbiol Infect Dis. 2010; 33:e127–e131.24. Sohal JS, Arsenault J, Labrecque O, Fairbrother JH, Roy JP, Fecteau G, L'Homme Y. Genetic structure of Mycobacterium avium subsp. paratuberculosis population in cattle herds in Quebec as revealed by using a combination of multilocus genomic analyses. J Clin Microbiol. 2014; 52:2764–2775.
Article25. Sonawane GG, Narnaware SD, Tripathi BN. Molecular epidemiology of Mycobacterium avium subspecies paratuberculosis in ruminants in different parts of India. Int J Mycobacteriol. 2016; 5:59–65.
Article26. Stevenson K, Alvarez J, Bakker D, Biet F, de Juan L, Denham S, Dimareli Z, Dohmann K, Gerlach GF, Heron I, Kopecna M, May L, Pavlik I, Sharp JM, Thibault VC, Willemsen P, Zadoks RN, Greig A. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol. 2009; 9:212.
Article27. Sweeney RW. Transmission of paratuberculosis. Vet Clin North Am Food Anim Pract. 1996; 12:305–312.
Article28. Thibault VC, Grayon M, Boschiroli ML, Hubbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol. 2007; 45:2404–2410.
Article29. Thibault VC, Grayon M, Boschiroli ML, Willery E, Allix-Béguec C, Stevenson K, Biet F, Supply P. Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates. J Clin Microbiol. 2008; 46:4091–4094.
Article30. Whittington RJ, Marsh IB, Whitlock RH. Typing of IS 1311 polymorphisms confirms that bison (Bison bison) with paratuberculosis in Montana are infected with a strain of Mycobacterium avium subsp. paratuberculosis distinct from that occurring in cattle and other domesticated livestock. Mol Cell Probes. 2001; 15:139–145.
Article31. Yoo HS, Shin SJ. Recent research on bovine paratuberculosis in South Korea. Vet Immunol Immunopathol. 2012; 148:23–28.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Effective DNA extraction method to improve detection of Mycobacterium avium subsp. paratuberculosis in bovine feces
- Tactics of Mycobacterium avium subsp. paratuberculosis for intracellular survival in mononuclear phagocytes
- Development of vaccines to Mycobacterium avium subsp. paratuberculosis infection
- Immunohistochemical localization of galectin-3 in the granulomatous lesions of paratuberculosis-infected bovine intestine
- Rapid visual detection of Mycobacterium avium subsp. paratuberculosis by recombinase polymerase amplification combined with a lateral flow dipstick