J Pathol Transl Med.
2018 May;52(3):148-156. 10.4132/jptm.2018.03.12.
Molecular Screening of Small Biopsy Samples Using Next-Generation Sequencing in Korean Patients with Advanced Non-small Cell Lung Cancer: Korean Lung Cancer Consortium (KLCC-13-01)
- Affiliations
-
- 1Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 2Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. silk.ahn@samsung.com
- 3CHA Bundang Medical Center, CHA University, Seongnam, Korea.
- 4Division of Medical Oncology, Yonsei Cancer Center, Seoul, Korea.
- 5Division of Hematology and Medical Oncology, Department of Internal Medicine, Gachon University Gil Medical Center, Incheon, Korea.
- 6Division of Oncology, Department of Hematology and Oncology, Ulsan University Hospital, Ulsan, Korea.
- 7Division of Medical Oncology, Chungbuk National University Hospital, Chungbuk National University College of Medicine, Cheongju, Korea.
- 8Department of Radiology and Center for Imaging Science, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 9Division of Pulmonary and Critical Care Medicine, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. kj2011.lee@samsung.com
- KMID: 2412926
- DOI: http://doi.org/10.4132/jptm.2018.03.12
Abstract
- BACKGROUND
Non-small cell lung cancer (NSCLC) is a common type of cancer with poor prognosis. As individual cancers exhibit unique mutation patterns, identifying and characterizing gene mutations in NSCLC might help predict patient outcomes and guide treatment. The aim of this study was to evaluate the clinical adequacy of molecular testing using next-generation sequencing (NGS) for small biopsy samples and characterize the mutational landscape of Korean patients with advanced NSCLC.
METHODS
DNA was extracted from small biopsy samples of 162 patients with advanced NSCLC. Targeted NGS of genomic alterations was conducted using Ion AmpliSeq Cancer Hotspot Panel v2.
RESULTS
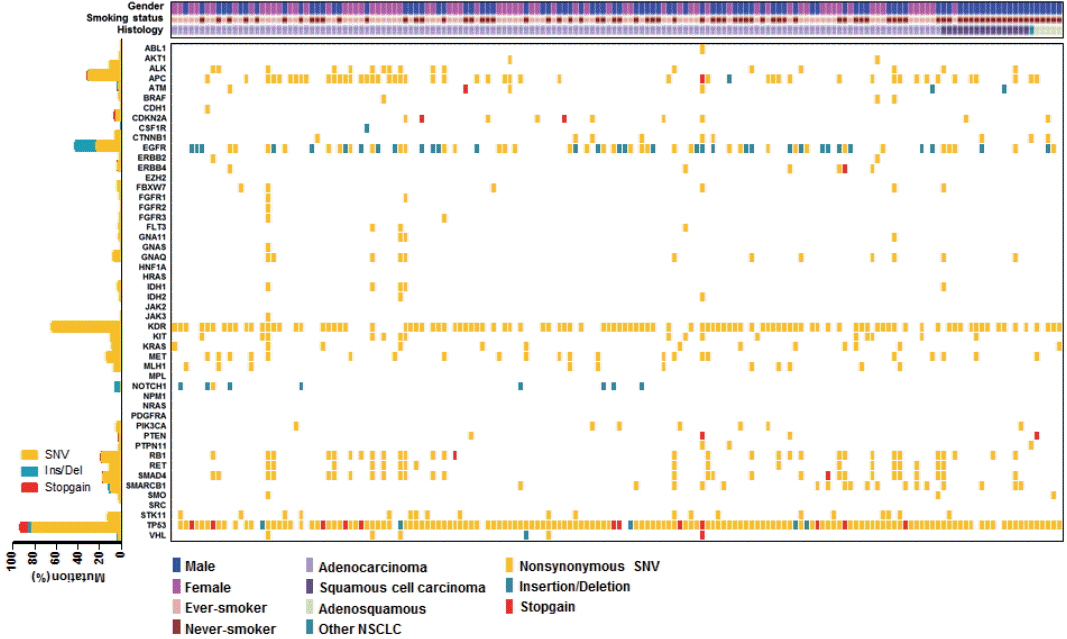
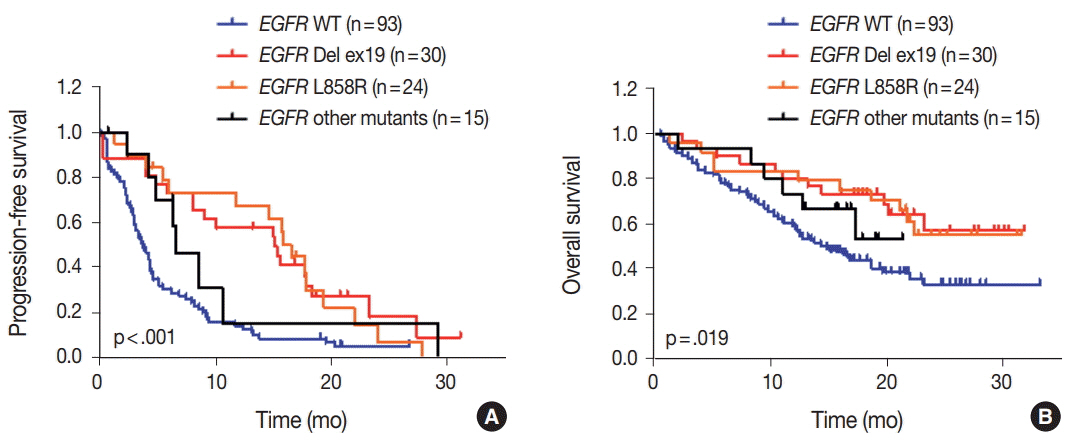
The median age of patients was 64 years (range, 32 to 83 years) and the majority had stage IV NSCLC at the time of cancer diagnosis (90%). Among the 162 patients, 161 patients (99.4%) had novel or hotspot mutations (range, 1 to 21 mutated genes). Mutations were found in 41 genes. Three of the most frequently mutated genes were TP53 (151, 93.2%), KDR (104, 64.2%), and epidermal growth factor receptor (EGFR; 69, 42.6%). We also observed coexistence of EGFR and other oncogene (such as KRAS, PIC3CA, PTEN, and STK11) mutations. Given that 69.6% (48/69) of EGFR mutant patients were treated with EGFR tyrosine kinase inhibitors, EGFR mutant status had higher prognostic ability in this study.
CONCLUSIONS
These results suggest that targeted NGS using small biopsy samples is feasible and allows for the detection of both common and rare mutations in NSCLC.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
J Pathol Transl Med. 2021;55(3):181-191. doi: 10.4132/jptm.2021.03.23.Landscape of
EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
J Pathol Transl Med. 2022;56(5):249-259. doi: 10.4132/jptm.2022.06.11.
Reference
-
1. Jung KW, Won YJ, Oh CM, et al. Prediction of cancer incidence and mortality in Korea, 2016. Cancer Res Treat. 2016; 48:451–7.
Article2. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014; 14:535–46.
Article3. Richer AL, Friel JM, Carson VM, Inge LJ, Whitsett TG. Genomic profiling toward precision medicine in non-small cell lung cancer: getting beyond EGFR. Pharmgenomics Pers Med. 2015; 8:63–79.4. Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014; 11:473–81.
Article5. Han JY, Kim SH, Lee YS, et al. Comparison of targeted next-generation sequencing with conventional sequencing for predicting the responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy in never-smokers with lung adenocarcinoma. Lung Cancer. 2014; 85:161–7.
Article6. Rathi V, Wright G, Constantin D, et al. Clinical validation of the 50 gene AmpliSeq Cancer Panel V2 for use on a next generation sequencing platform using formalin fixed, paraffin embedded and fine needle aspiration tumour specimens. Pathology. 2017; 49:75–82.
Article7. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014; 311:1998–2006.
Article8. Ballester LY, Luthra R, Kanagal-Shamanna R, Singh RR. Advances in clinical next-generation sequencing: target enrichment and sequencing technologies. Expert Rev Mol Diagn. 2016; 16:357–72.
Article9. Padmanabhan V, Steinmetz HB, Rizzo EJ, et al. Improving adequacy of small biopsy and fine-needle aspiration specimens for molecular testing by next-generation sequencing in patients with lung cancer: a quality improvement study at Dartmouth-Hitchcock Medical Center. Arch Pathol Lab Med. 2017; 141:402–9.
Article10. Zheng G, Tsai H, Tseng LH, et al. Test feasibility of next-generation sequencing assays in clinical mutation detection of small biopsy and fine needle aspiration specimens. Am J Clin Pathol. 2016; 145:696–702.
Article11. Kim ST, Lee J, Hong M, et al. The NEXT-1 (Next generation pErsonalized tX with mulTi-omics and preclinical model) trial: prospective molecular screening trial of metastatic solid cancer patients, a feasibility analysis. Oncotarget. 2015; 6:33358–68.
Article12. Kim HJ, Lee KY, Kim YC, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012; 75:321–5.
Article13. Ku BM, Jung HA, Sun JM, et al. High-throughput profiling identifies clinically actionable mutations in salivary duct carcinoma. J Transl Med. 2014; 12:299.
Article14. Lee GD, Lee SE, Oh DY, et al. MET exon 14 skipping mutations in lung adenocarcinoma: clinicopathologic implications and prognostic Values. J Thorac Oncol. 2017; 12:1233–46.15. Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer. 2013; 14:205–14.16. Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012; 13:239–46.17. Kuan FC, Kuo LT, Chen MC, et al. Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: a systematic review and meta-analysis. Br J Cancer. 2015; 113:1519–28.18. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–50.19. Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol. 2013; 24:2371–6.
Article20. Lim SM, Kim HR, Cho EK, et al. Targeted sequencing identifies genetic alterations that confer primary resistance to EGFR tyrosine kinase inhibitor (Korean Lung Cancer Consortium). Oncotarget. 2016; 7:36311–20.
Article21. Mäki-Nevala S, Sarhadi VK, Rönty M, et al. Hot spot mutations in Finnish non-small cell lung cancers. Lung Cancer. 2016; 99:102–10.
Article22. Murakami I, Hiyama K, Ishioka S, Yamakido M, Kasagi F, Yokosaki Y. p53 gene mutations are associated with shortened survival in patients with advanced non-small cell lung cancer: an analysis of medically managed patients. Clin Cancer Res. 2000; 6:526–30.23. Lim EH, Zhang SL, Li JL, et al. Using whole genome amplification (WGA) of low-volume biopsies to assess the prognostic role of EGFR, KRAS, p53, and CMET mutations in advanced-stage nonsmall cell lung cancer (NSCLC). J Thorac Oncol. 2009; 4:12–21.24. Glubb DM, Cerri E, Giese A, et al. Novel functional germline variants in the VEGF receptor 2 gene and their effect on gene expression and microvessel density in lung cancer. Clin Cancer Res. 2011; 17:5257–67.
Article25. Silva IP, Salhi A, Giles KM, et al. Identification of a novel pathogenic germline KDR variant in melanoma. Clin Cancer Res. 2016; 22:2377–85.
Article26. Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol. 2013; 31:1112–21.27. Kim HR, Cho BC, Shim HS, et al. Prediction for response duration to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma. Lung Cancer. 2014; 83:374–82.
Article28. Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, et al. Different prognostic impact of STK11 mutations in non-squamous non-small-cell lung cancer. Oncotarget. 2017; 8:23831–40.29. Gleeson FC, Kipp BR, Levy MJ, et al. Somatic STK11 and concomitant STK11/KRAS mutational frequency in stage IV lung adenocarcinoma adrenal metastases. J Thorac Oncol. 2015; 10:531–4.30. Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012; 150:1107–20.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecularly Targeted Therapy for Lung Cancer : Recent Topics
- Recent advances in diagnostic technologies in lung cancer
- The Utilization of Cytologic Fine-Needle Aspirates of Lung Cancer for Molecular Diagnostic Testing
- Molecular Pathogenesis of Non-Small Cell Lung Carcinomas
- A Case of Spontaneous Regression of Small Cell Lung Cancer