J Lung Cancer.
2012 Jun;11(1):12-20. 10.6058/jlc.2012.11.1.12.
Molecular Pathogenesis of Non-Small Cell Lung Carcinomas
- Affiliations
-
- 1Departments of Pathology and Thoracic/Head and Neck Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA. iiwistuba@mdanderson.org
- KMID: 2199871
- DOI: http://doi.org/10.6058/jlc.2012.11.1.12
Abstract
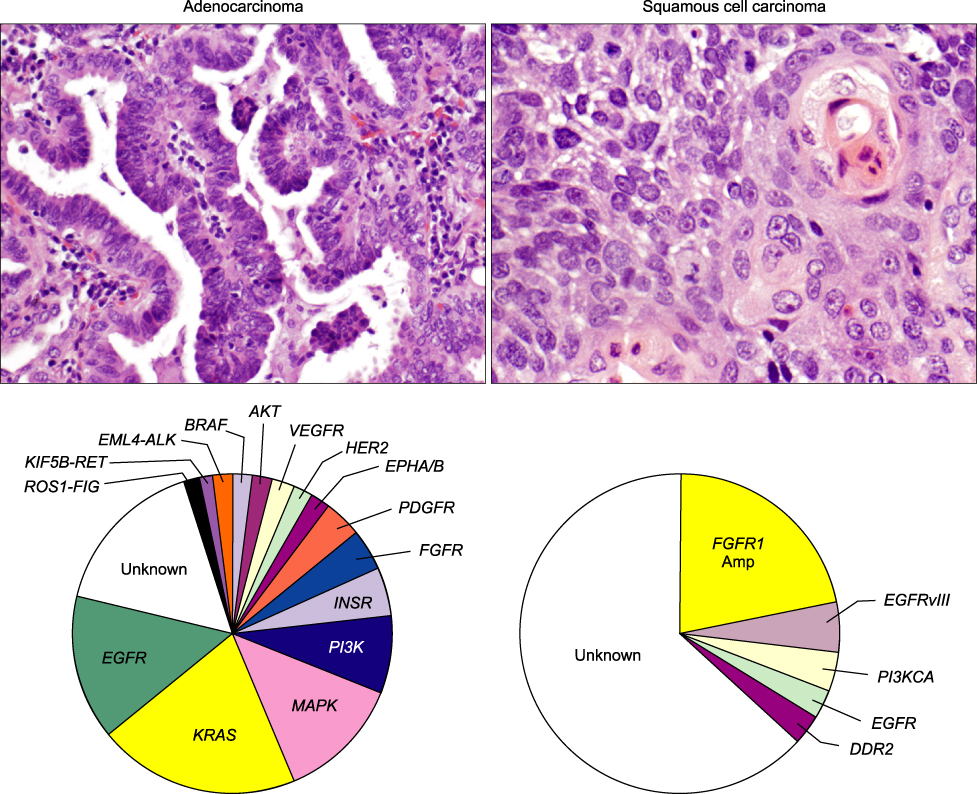
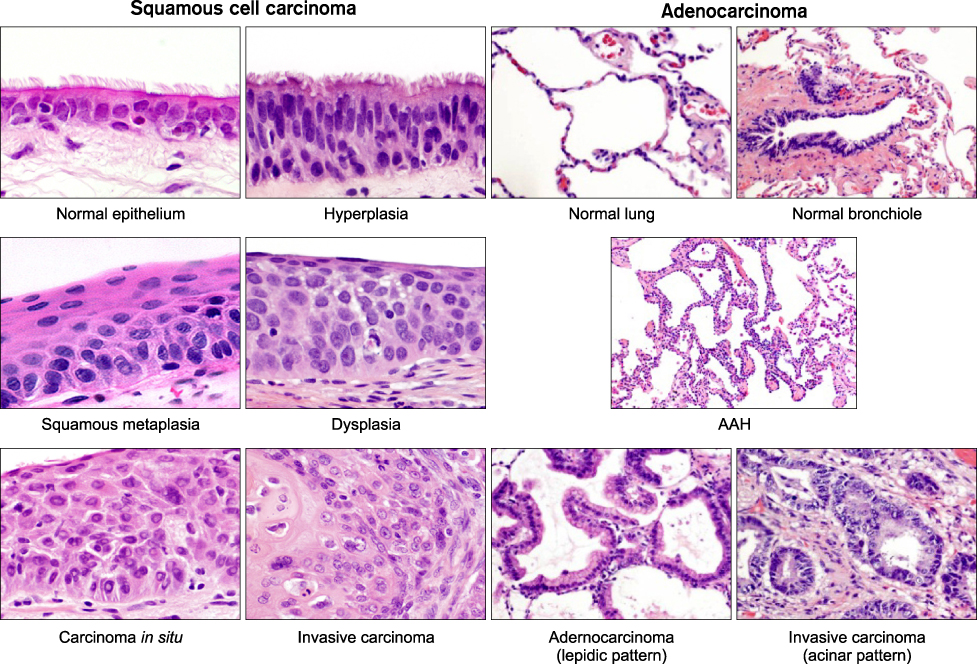
- Non-small cell lung carcinoma (NSCLC) from histological and biological perspectives is a complex neoplasm. The sequential preneoplastic changes have been defined for centrally arising squamous cell carcinomas (SCCs) of the lung, and they are less documented for peripherally arising adenocarcinomas. The main morphologic forms of preneoplastic lesions recognized in the lung are squamous dysplasias for SCC, and atypical adenomatous hyperplasia for adenocarcinoma. Several studies have provided information regarding the molecular characterization of lung preneoplastic changes, especially for SCC. These molecular changes have been detected in the histologically normal and abnormal respiratory epithelium of smokers and patients with lung cancer, phenomenon known as field of cancerization. Our improved understanding of the changes and origins of the field of cancerization can be applied clinically to improve early detection of lung cancer. In the last decade, significant progress has been made in the characterization of molecular abnormalities in NSCLC tumors that are being used as molecular targets and predictive biomarkers for patients' selection for targeted therapy. As our understanding of the biology of the molecular pathogenesis of lung cancer evolves, there is an opportunity to use this knowledge for the development of novel chemoprevention strategies using those molecularly targeted agents used to treat advanced lung cancer, a concept coined as reverse migration. The rapid development of technologies for large-scale molecular analysis, includeing microarrays and next-generation sequencing will facilitate high-throughput molecular analysis of lung cancer preneoplastic lesions and the field of cancerization.
Keyword
MeSH Terms
Figure
Reference
-
1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010. 60:277–300.
Article2. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008. 359:1367–1380.
Article3. Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011. 32:22–31.
Article4. Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011. 12:175–180.
Article5. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011. 144:646–674.
Article6. Kadara H, Behrens C, Yuan P, et al. A five-gene and corresponding protein signature for stage-I lung adenocarcinoma prognosis. Clin Cancer Res. 2011. 17:1490–1501.
Article7. Gold KA, Wistuba II, Kim ES. New strategies in squamous cell carcinoma of the lung: identification of tumor drivers to personalize therapy. Clin Cancer Res. 2012. 18:3002–3007.
Article8. Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012. 18:2443–2451.
Article9. Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009. 41:1238–1242.
Article10. Yuan P, Kadara H, Behrens C, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010. 5:e9112.
Article11. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010. 2:62ra93.12. Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011. 1:78–89.13. Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011. 6:e20351.
Article14. Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC. World Health Organization classification of tumours: pathology and genetics of tumours of the lung, pleura, thymus and heart. 2004. Lyon: IARC Press;9–124.15. Solis LM, Behrens C, Raso MG, et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer. 2012. 118:2889–2899.
Article16. Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008. 455:1069–1075.17. Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012. 18:375–377.
Article18. Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012. 18:378–381.
Article19. Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012. 18:382–384.
Article20. Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012. 30:863–870.
Article21. Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006. 118:257–262.
Article22. Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005. 97:339–346.
Article23. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009. 361:947–957.
Article24. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007. 448:561–566.
Article25. Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009. 22:508–515.
Article26. Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011. 12:1004–1012.
Article27. Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008. 3:13–17.
Article28. Dacic S, Shuai Y, Yousem S, Ohori P, Nikiforova M. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol. 2010. 23:159–168.
Article29. Siegfried JM, Gillespie AT, Mera R, et al. Prognostic value of specific KRAS mutations in lung adenocarcinomas. Cancer Epidemiol Biomarkers Prev. 1997. 6:841–847.30. Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008. 14:5731–5734.
Article31. Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007. 13:2890–2896.
Article32. Tang X, Kadara H, Behrens C, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011. 17:2434–2443.
Article33. Whitsett J. A lungful of transcription factors. Nat Genet. 1998. 20:7–8.
Article34. Perner S, Wagner PL, Soltermann A, et al. TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J Pathol. 2009. 217:65–72.
Article35. Kerr KM. Pulmonary preinvasive neoplasia. J Clin Pathol. 2001. 54:257–271.
Article36. Westra WH. Early glandular neoplasia of the lung. Respir Res. 2000. 1:163–169.
Article37. Wistuba II. Genetics of preneoplasia: lessons from lung cancer. Curr Mol Med. 2007. 7:3–14.
Article38. Park IW, Wistuba II, Maitra A, et al. Multiple clonal abnormalities in the bronchial epithelium of patients with lung cancer. J Natl Cancer Inst. 1999. 91:1863–1868.
Article39. Wistuba II, Mao L, Gazdar AF. Smoking molecular damage in bronchial epithelium. Oncogene. 2002. 21:7298–7306.
Article40. McCaughan F, Pole JC, Bankier AT, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med. 2010. 182:83–91.
Article41. McCaughan F, Pipinikas CP, Janes SM, George PJ, Rabbitts PH, Dear PH. Genomic evidence of pre-invasive clonal expansion, dispersal and progression in bronchial dysplasia. J Pathol. 2011. 224:153–159.
Article42. Wistuba II. Histologic evaluation of bronchial squamous lesions: any role in lung cancer risk assessment? Clin Cancer Res. 2005. 11:1358–1360.
Article43. Colby TV, Wistuba II, Gazdar A. Precursors to pulmonary neoplasia. Adv Anat Pathol. 1998. 5:205–215.
Article44. Kitamura H, Kameda Y, Ito T, Hayashi H. Atypical adenomatous hyperplasia of the lung. Implications for the pathogenesis of peripheral lung adenocarcinoma. Am J Clin Pathol. 1999. 111:610–622.
Article45. Osanai M, Igarashi T, Yoshida Y. Unique cellular features in atypical adenomatous hyperplasia of the lung: ultrastructural evidence of its cytodifferentiation. Ultrastruct Pathol. 2001. 25:367–373.
Article46. Westra WH, Baas IO, Hruban RH, et al. K-ras oncogene activation in atypical alveolar hyperplasias of the human lung. Cancer Res. 1996. 56:2224–2228.47. Tominaga M, Sueoka N, Irie K, et al. Detection and discrimination of preneoplastic and early stages of lung adenocarcinoma using hnRNP B1 combined with the cell cycle-related markers p16, cyclin D1, and Ki-67. Lung Cancer. 2003. 40:45–53.
Article48. Nakanishi K, Kawai T, Kumaki F, Hiroi S, Mukai M, Ikeda E. Survivin expression in atypical adenomatous hyperplasia of the lung. Am J Clin Pathol. 2003. 120:712–719.
Article49. Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol. 2005. 29:633–639.
Article50. Tang X, Shigematsu H, Bekele BN, et al. EGFR tyrosine kinase domain mutations are detected in histologically normal respiratory epithelium in lung cancer patients. Cancer Res. 2005. 65:7568–7572.
Article51. Tang X, Varella-Garcia M, Xavier AC, et al. Epidermal growth factor receptor abnormalities in the pathogenesis and progression of lung adenocarcinomas. Cancer Prev Res (Phila). 2008. 1:192–200.
Article52. Rawlins EL, Okubo T, Que J, et al. Epithelial stem/progenitor cells in lung postnatal growth, maintenance, and repair. Cold Spring Harb Symp Quant Biol. 2008. 73:291–295.
Article53. Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005. 121:823–835.
Article54. Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008. 15:504–514.
Article55. Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010. 70:9937–9948.
Article56. Kadara H, Wistuba II. Field cancerization in non-small cell lung cancer: implications in disease pathogenesis. Proc Am Thorac Soc. 2012. 9:38–42.57. Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953. 6:963–968.
Article58. Nelson MA, Wymer J, Clements N Jr. Detection of K-ras gene mutations in non-neoplastic lung tissue and lung cancers. Cancer Lett. 1996. 103:115–121.
Article59. Spira A, Beane J, Shah V, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004. 101:10143–10148.
Article60. Spira A, Beane JE, Shah V, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007. 13:361–366.
Article61. Gold KA, Kim ES, Lee JJ, Wistuba II, Farhangfar CJ, Hong WK. The BATTLE to personalize lung cancer prevention through reverse migration. Cancer Prev Res (Phila). 2011. 4:962–972.
Article62. Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010. 11:685–696.
Article63. Beane J, Vick J, Schembri F, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila). 2011. 4:803–817.
Article