J Vet Sci.
2018 Jan;19(1):35-43. 10.4142/jvs.2018.19.1.35.
Analysis of structure-function relationship in porcine rotavirus A enterotoxin gene
- Affiliations
-
- 1Indian Council of Agricultural Research-Indian Veterinary Research Institute, Uttar Pradesh 243122, India. malikyps@ivri.res.in
- 2Department of Biomedical Sciences, One Health Center for Zoonoses and Tropical Veterinary Medicine, Ross University School of Veterinary Medicine, P.O. Box 334, Basseterre, Federation of Saint Kitts and Nevis.
- 3Sapporo Medical University School of Medicine, Sapporo 060-8556, Japan.
- 4Institute for Veterinary Medical Research, Centre for Agricultural Research, Hungarian Academy of Sciences, Budapest 1143, Hungary.
- KMID: 2402693
- DOI: http://doi.org/10.4142/jvs.2018.19.1.35
Abstract
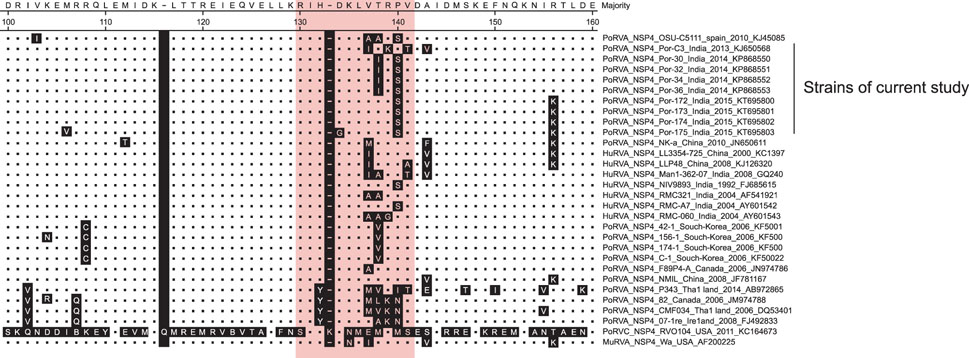
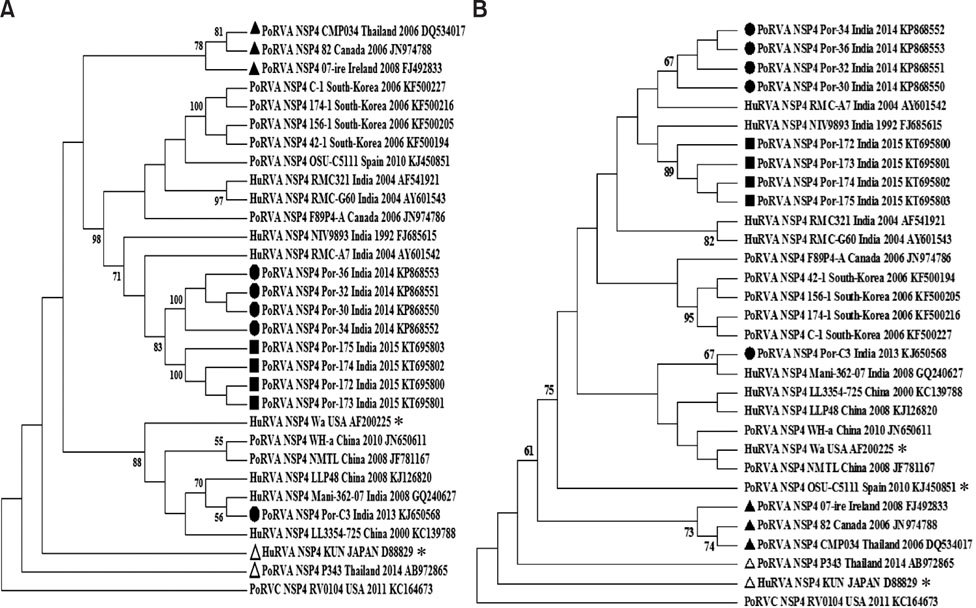
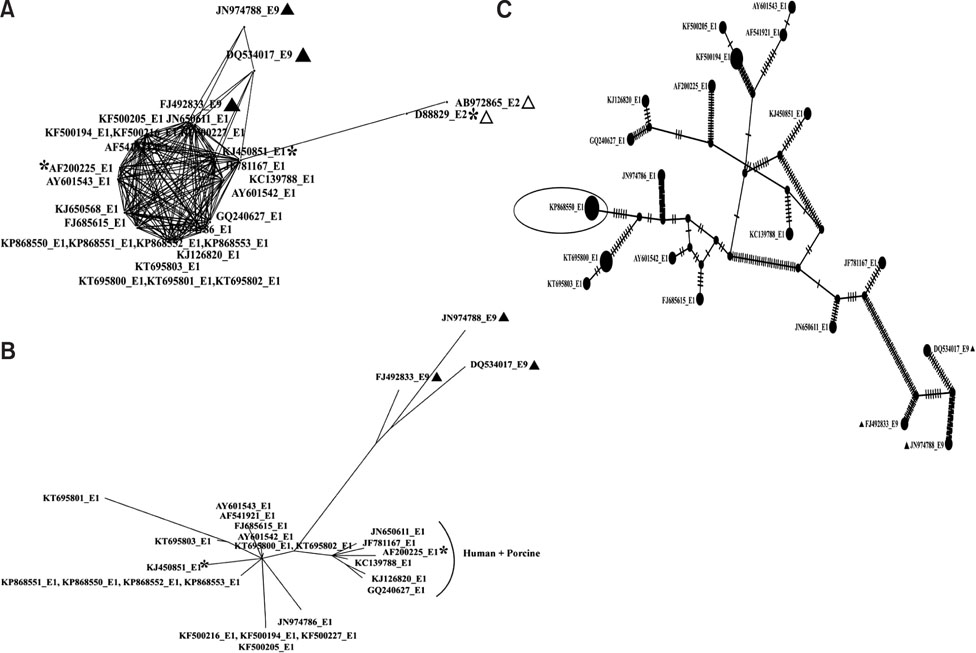
- Rotavirus (RV)-infected piglets are presumed to be latent sources of heterologous RV infection in humans and other animals. In RVs, non-structural protein 4 (NSP4) is the major virulence factor with pleiotropic properties. In this study, we analyzed the nsp4 gene from porcine RVs isolated from diarrheic and non-diarrheic cases at different levels of protein folding to explore correlations to diarrhea-inducing capabilities and evolution of nsp4 in the porcine population. Full-length nsp4 genes were amplified, cloned, sequenced, and then analyzed for antigenic epitopes, RotaC classification, homology, genetic relationship, modeling of NSP4 protein, and prediction of post-translational modification. RV presence was observed in both diarrheic and non-diarrheic piglets. All nsp4 genes possessed the E1 genotype. Comparison of primary, secondary, and tertiary structure and the prediction of post-translational modifications of NSP4 from diarrheic and non-diarrheic piglets revealed no apparent differences. Sequence analysis indicated that nsp4 genes have a multi-phyletic evolutionary origin and exhibit species independent genetic diversity. The results emphasize the evolution of the E9 nsp4 genotype from the E1 genotype and suggest that the diarrhea-inducing capability of porcine RVs may not be exclusively linked to its enterotoxin gene.
MeSH Terms
Figure
Reference
-
1. Angel J, Tang B, Feng N, Greenberg HB, Bass D. Studies of the role for NSP4 in the pathogenesis of homologous murine rotavirus diarrhea. J Infect Dis. 1998; 177:455–458.
Article2. Attoui H, Becnel J, Belaganahalli S, Bergoin M, Brussaard C, Chappell J, Ciarlet M, del Vas M, Dermody T, Dormitzer P. Family Reoviridae. In : King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Classification and Nomenclature of Viruses. Waltham: Academic Press;2012. p. 541–637.3. Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006; 80:6061–6071.
Article4. Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999; 294:1351–1362.
Article5. Bora DP, Barman NN, Bhattacharyya DK, Dutta LJ. Faecal excretion of rotavirus by naturally infected pregnant sows in organized pig farms–a potent source of infection to newborns. Indian J Anim Sci. 2011; 81:575–577.6. Campagna M, Marcos-Villar L, Arnoldi F, de la Cruz-Herrera CF, Gallego P, González-Santamaría J, González D, Lopitz-Otsoa F, Rodriguez MS, Burrone OR, Rivas C. Rotavirus viroplasm proteins interact with the cellular SUMOylation system: implications for viroplasm-like structure formation. J Virol. 2013; 87:807–817.
Article7. Chang KO, Kim YJ, Saif LJ. Comparisons of nucleotide and deduced amino acid sequences of NSP4 genes of virulent and attenuated pairs of group A and C rotaviruses. Virus Genes. 1999; 18:229–233.8. Chattopadhyay S, Bagchi P, Dutta D, Mukherjee A, Kobayashi N, Chawlasarkar M. Computational identification of posttranslational modification sites and functional families reveal possible moonlighting role of rotaviral proteins. Bioinformation. 2010; 4:448–451.
Article9. Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000; 9:1657–1659.
Article10. Deepa R, Rao CD, Suguna K. Structure of the extended diarrhea-inducing domain of rotavirus enterotoxigenic protein NSP4. Arch Virol. 2007; 152:847–859.
Article11. Dhama K, Chauhan RS, Mahendran M, Malik SVS. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun. 2009; 33:1–23.
Article12. Dubal ZB, Bhilegaonkar KN, Barbuddhe SB, Kolhe RP, Kaur S, Rawat S, Nambiar P, Karunakaran M. Prevalence and genotypic (G and P) determination of porcine group A rotaviruses from different regions of India. Trop Anim Health Prod. 2013; 45:609–615.
Article13. Estes MK, Kapikian AZ. Rotaviruses. In : Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Fields Virology. 5 ed. Vol. 2. Philadelphia: Lippincott Williams & Wilkins;2007. p. 1917–1974.14. Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995; 11:681–684.
Article15. Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006; 23:254–267.
Article16. Jagannath MR, Vethanayagam RR, Reddy BS, Raman S, Rao CD. Characterization of human symptomatic rotavirus isolates MP409 and MP480 having ‘long’ RNA electropherotype and subgroup I specificity, highly related to the P6[1],G8 type bovine rotavirus A5, from Mysore, India. Arch Virol. 2000; 145:1339–1357.
Article17. Komoto S, Pongsuwanna Y, Ide T, Wakuda M, Guntapong R, Dennis FE, Haga K, Fujii Y, Katayama K, Taniguchi K. Whole genomic analysis of porcine G10P[5] rotavirus strain P343 provides evidence for bovine-to-porcine interspecies transmission. Vet Microbiol. 2014; 174:577–583.
Article18. Kuga K, Miyazaki A, Suzuki T, Takagi M, Hattori N, Katsuda K, Mase M, Sugiyama M, Tsunemitsu H. Genetic diversity and classification of the outer capsid glycoprotein VP7 of porcine group B rotaviruses. Arch Virol. 2009; 154:1785–1795.
Article19. Kumar M, Bhilegaonkar KN, Agarwal RK. Prevalence and characterization of rotavirus from faecal samples of children and animals. Indian J Anim Sci. 2011; 81:993–999.20. Kusumakar AL, Savita , Malik YPS, Minakshi , Prasad G. Occurrence of mammalian group A rotavirus in swine population of central India. Indian J Anim Sci. 2008; 78:475–477.21. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25:1451–1452.
Article22. Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009; 9:238.
Article23. Malik YS, Kumar N, Sharma K, Ghosh S, Bányai K, Balasubramanian G, Kobayashi N, Matthijnssens J. Molecular analysis of non structural rotavirus group A enterotoxin gene of bovine origin from India. Infect Genet Evol. 2014; 25:20–27.
Article24. Malik YS, Kumar N, Sharma K, Haq AA, Kumar A, Prasad M. Sequence and phylogenetic analysis of bovine rotavirus isolates (G6 genotypes) from India. Adv Anim Vet Sci. 2013; 1:41–43.25. Malik YS, Kumar N, Sharma K, Sircar S, Dhama K, Bora DP, Dutta TK, Prasad M, Tiwari AK. Rotavirus diarrhea in piglets: a review on epidemiology, genetic diversity and zoonotic risks. Indian J Anim Sci. 2014; 84:1035–1042.26. Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Bányai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gómara M, Johne R, Kirkwood CD, Martella V, Mertens PPC, Nakagomi O, Parreño V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol. 2011; 156:1397–1413.
Article27. Matthijnssens J, Rahman M, Ciarlet M, Zeller M, Heylen E, Nakagomi T, Uchida R, Hassan Z, Azim T, Nakagomi O, Van Ranst M. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerg Infect Dis. 2010; 16:625–630.
Article28. Médici KC, Barry AF, Alfieri AF, Alfieri AA. Porcine rotavirus groups A, B, and C identified by polymerase chain reaction in a fecal sample collection with inconclusive results by polyacrylamide gel electrophoresis. J Swine Health Prod. 2011; 19:146–150.29. Miyazaki A, Kuga K, Suzuki T, Kohmoto M, Katsuda K, Tsunemitsu H. Annual changes in predominant genotypes of rotavirus A detected in the feces of pigs in various developmental stages raised on a conventional farm. Vet Microbiol. 2013; 163:162–166.
Article30. Mukherjee A, Ghosh S, Bagchi P, Dutta D, Chattopadhyay S, Kobayashi N, Chawla-Sarkar M. Full genomic analyses of human rotavirus G4P[4], G4P[6], G9P[19] and G10P[6] strains from North-eastern India: evidence for interspecies transmission and complex reassortment events. Clin Microbiol Infect. 2011; 17:1343–1346.
Article31. Oka T, Nakagomi T, Nakagomi O. A Lack of consistent amino acid substitutions in NSP4 between rotaviruses derived from diarrheal and asymptomaticallyXMLLink_XYZinfected kittens. Microbiol Immunol. 2001; 45:173–177.
Article32. Papp H, Al-Mutairi L, Chehadeh W, Farkas S, Lengyel G, Jakab F, Martella V, Szücs G, Bányai K. Novel NSP4 genotype in a camel G10P[15] rotavirus strain. Acta Microbiol Immunol Hung. 2012; 59:411–421.
Article33. Papp H, László B, Jakab F, Ganesh B, De Grazia S, Matthijnssens J, Ciarlet M, Martella V, Bányai K. Review of group A rotavirus strains reported in swine and cattle. Vet Microbiol. 2013; 165:190–199.
Article34. Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998; 14:817–818.
Article35. Rajasekaran D, Sastri NP, Marathahalli JR, Indi SS, Pamidimukkala K, Suguna K, Rao CD. The flexible C terminus of the rotavirus non-structural protein NSP4 is an important determinant of its biological properties. J Gen Virol. 2008; 89:1485–1496.
Article36. Srivastava S, Jain A. Rotavirus nonstructural protein 4 (NSP4)-viral enterotoxin with multiple roles in pathogenesis of diarrhoea in children. J Appl Pharm Sci. 2015; 5:146–153.
Article37. Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. Precision mapping of the human O–GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013; 32:1478–1488.
Article38. Suguna K, Rao CD. Rotavirus nonstructural proteins: a structural perspective. Curr Sci. 2010; 98:352–359.39. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30:2725–2729.
Article40. Tate JE, Chitambar S, Esposito DH, Sarkar R, Gladstone B, Ramani S, Raghava MV, Sowmyanarayanan TV, Gandhe S, Arora R, Rarashar UD, Kang G. Disease and economic burden of rotavirus diarrhoea in India. Vaccine. 2009; 27:Suppl 5. F18–F24.
Article41. Theuns S, Heylen E, Zeller M, Roukaerts IDM, Desmarets LMB, Van Ranst M, Nauwynck HJ, Matthijnssens J. Complete genome characterization of recent and ancient Belgian pig group A rotaviruses and assessment of their evolutionary relationship with human rotaviruses. J Virol. 2015; 89:1043–1057.
Article42. Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015; 12:7–8.
Article43. Yang W, McCrae MA. The molecular biology of rotaviruses X: intercellular dissemination of rotavirus NSP4 requires glycosylation and is mediated by direct cell-cell contact through cytoplasmic extrusions. Arch Virol. 2012; 157:305–314.
Article44. Zhang M, Zeng CQY, Dong Y, Ball JM, Saif LJ, Morris AP, Estes MK. Mutations in rotavirus nonstructural glycoprotein NSP4 are associated with altered virus virulence. J Virol. 1998; 72:3666–3672.
Article45. Zhang M, Zeng CQY, Morris AP, Estes MK. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J Virol. 2000; 74:11663–11670.
Article46. Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014; 42:W325–W330.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Cloning and Sequencing of Nonstructural Protein (NSP4) Gene of Bovine Rotavirus Isolated in Korea
- Role of Rotavirus Enterotoxin NSP4 in the Inflammatory Response in Murine Macrophage RAW 264.7 Cells
- Rotavirus Vaccine
- Rotavirus Vaccines
- The laboratory test procedure to confirm rotavirus vaccine infection in severe complex immunodeficiency patients