Immune Netw.
2017 Oct;17(5):298-306. 10.4110/in.2017.17.5.298.
Current Understanding in Neutrophil Differentiation and Heterogeneity
- Affiliations
-
- 1Department of Physiology, College of Medicine, Kyungpook National University, Daegu 41944, Korea. cwhong@knu.ac.kr
- KMID: 2400635
- DOI: http://doi.org/10.4110/in.2017.17.5.298
Abstract
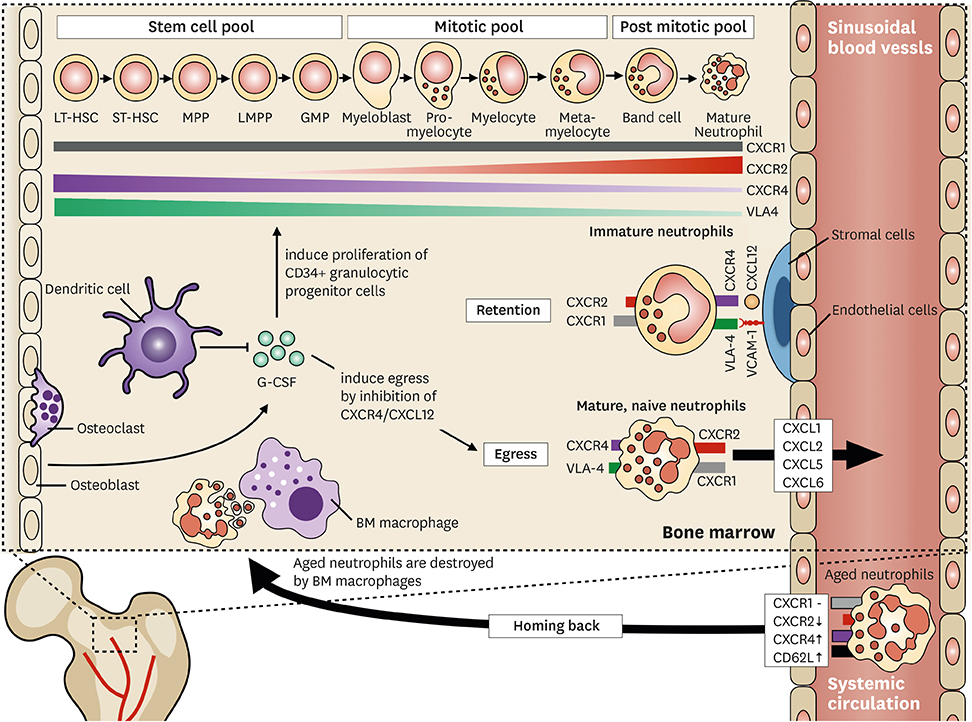
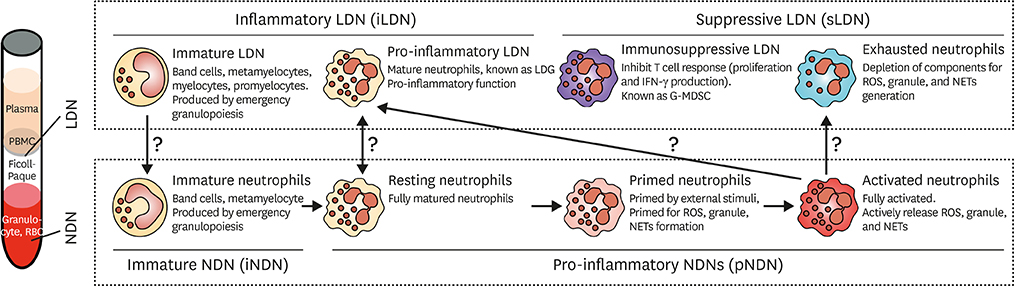
- Neutrophils are professional phagocytes that conduct effectors functions in the innate immune systems. They are differentiated in the bone marrow (BM) and terminally differentiated neutrophils are then released into systemic circulation. Neutrophils migrate into inflammatory foci through extravasation, reverse transmigration, and chemotaxis. As neutrophils arrive at a target site, they actively participate in eliminating pathogens. They phagocytose bacteria, and eliminate them through the generation of reactive oxygen species (ROS), release of protease-enriched granules, and formation of neutrophil extracellular traps (NETs). Since neutrophils are equipped with toxic arsenals, the activation of neutrophils is tightly controlled. Priming is the process of unlocking safety mechanisms before complete activation of neutrophils. Since the first discovery of neutrophils, they were considered as a homogeneous population with an inflammatory phenotype. However, heterogenous populations of neutrophils were discovered under physiological and pathological conditions. This review outlines the normal differentiation of neutrophils in the BM, and discusses the current understandings of neutrophil heterogeneity.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Aging and the Immune System: the Impact of Immunosenescence on Viral Infection, Immunity and Vaccine Immunogenicity
Soo-Jin Oh, Jae Kyung Lee, Ok Sarah Shin
Immune Netw. 2019;19(6):. doi: 10.4110/in.2019.19.e37.
Reference
-
1. Nauseef WM, Borregaard N. Neutrophils at work. Nat Immunol. 2014; 15:602–611.
Article2. El-Benna J, Hurtado-Nedelec M, Marzaioli V, Marie JC, Gougerot-Pocidalo MA, Dang PM. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunol Rev. 2016; 273:180–193.
Article3. Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006; 6:541–550.
Article4. Cowland JB, Borregaard N. Granulopoiesis and granules of human neutrophils. Immunol Rev. 2016; 273:11–28.
Article5. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004; 303:1532–1535.
Article6. Metchnikoff E. Immunity in Infective Diseases. Cambridge: Cambridge University Press;1905.7. Beyrau M, Bodkin JV, Nourshargh S. Neutrophil heterogeneity in health and disease: a revitalized avenue in inflammation and immunity. Open Biol. 2012; 2:120134.
Article8. Carmona-Rivera C, Kaplan MJ. Low-density granulocytes: a distinct class of neutrophils in systemic autoimmunity. Semin Immunopathol. 2013; 35:455–463.
Article9. Scapini P, Cassatella MA. Social networking of human neutrophils within the immune system. Blood. 2014; 124:710–719.
Article10. Sagiv JY, Michaeli J, Assi S, Mishalian I, Kisos H, Levy L, Damti P, Lumbroso D, Polyansky L, Sionov RV, et al. Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell Reports. 2015; 10:562–573.
Article11. Silvestre-Roig C, Hidalgo A, Soehnlein O. Neutrophil heterogeneity: implications for homeostasis and pathogenesis. Blood. 2016; 127:2173–2181.
Article12. Scapini P, Marini O, Tecchio C, Cassatella MA. Human neutrophils in the saga of cellular heterogeneity: insights and open questions. Immunol Rev. 2016; 273:48–60.
Article13. Görgens A, Radtke S, Möllmann M, Cross M, Dürig J, Horn PA, Giebel B. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Reports. 2013; 3:1539–1552.
Article14. Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016; 16:431–446.
Article15. Pruchniak MP, Arazna M, Demkow U. Life of neutrophil: from stem cell to neutrophil extracellular trap. Respir Physiol Neurobiol. 2013; 187:68–73.
Article16. Futosi K, Fodor S, Mócsai A. Reprint of Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol. 2013; 17:1185–1197.
Article17. Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. 2010; 31:318–324.
Article18. Elghetany MT. Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis. 2002; 28:260–274.
Article19. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014; 505:327–334.
Article20. Petty JM, Lenox CC, Weiss DJ, Poynter ME, Suratt BT. Crosstalk between CXCR4/stromal derived factor-1 and VLA-4/VCAM-1 pathways regulates neutrophil retention in the bone marrow. J Immunol. 2009; 182:604–612.
Article21. Eash KJ, Greenbaum AM, Gopalan PK, Link DC. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J Clin Invest. 2010; 120:2423–2431.
Article22. Rankin SM. The bone marrow: a site of neutrophil clearance. J Leukoc Biol. 2010; 88:241–251.
Article23. Sadik CD, Kim ND, Luster AD. Neutrophils cascading their way to inflammation. Trends Immunol. 2011; 32:452–460.
Article24. Semerad CL, Christopher MJ, Liu F, Short B, Simmons PJ, Winkler I, Levesque JP, Chappel J, Ross FP, Link DC. G-CSF potently inhibits osteoblast activity and CXCL12 mRNA expression in the bone marrow. Blood. 2005; 106:3020–3027.
Article25. Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014; 20:833–846.
Article26. Lord BI, Bronchud MH, Owens S, Chang J, Howell A, Souza L, Dexter TM. The kinetics of human granulopoiesis following treatment with granulocyte colony-stimulating factor in vivo . Proc Natl Acad Sci USA. 1989; 86:9499–9503.
Article27. Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002; 17:413–423.
Article28. Kim HK, De La Luz Sierra M, Williams CK, Gulino AV, Tosato G. G-CSF down-regulation of CXCR4 expression identified as a mechanism for mobilization of myeloid cells. Blood. 2006; 108:812–820.
Article29. Jiao J, Dragomir AC, Kocabayoglu P, Rahman AH, Chow A, Hashimoto D, Leboeuf M, Kraus T, Moran T, Carrasco-Avino G, et al. Central role of conventional dendritic cells in regulation of bone marrow release and survival of neutrophils. J Immunol. 2014; 192:3374–3382.
Article30. Duffy D, Perrin H, Abadie V, Benhabiles N, Boissonnas A, Liard C, Descours B, Reboulleau D, Bonduelle O, Verrier B, et al. Neutrophils transport antigen from the dermis to the bone marrow, initiating a source of memory CD8+ T cells. Immunity. 2012; 37:917–929.
Article31. Gallin JI. Human neutrophil heterogeneity exists, but is it meaningful? Blood. 1984; 63:977–983.
Article32. Mishalian I, Granot Z, Fridlender ZG. The diversity of circulating neutrophils in cancer. Immunobiology. 2017; 222:82–88.
Article33. Hacbarth E, Kajdacsy-Balla A. Low density neutrophils in patients with systemic lupus erythematosus, rheumatoid arthritis, and acute rheumatic fever. Arthritis Rheum. 1986; 29:1334–1342.
Article34. Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014; 14:302–314.
Article35. Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006; 7:732–739.36. Levin R, Grinstein S, Canton J. The life cycle of phagosomes: formation, maturation, and resolution. Immunol Rev. 2016; 273:156–179.
Article37. Abdel-Latif D, Steward M, Macdonald DL, Francis GA, Dinauer MC, Lacy P. Rac2 is critical for neutrophil primary granule exocytosis. Blood. 2004; 104:832–839.
Article38. Ward PA. The dark side of C5a in sepsis. Nat Rev Immunol. 2004; 4:133–142.
Article39. Lacy P. Mechanisms of degranulation in neutrophils. Allergy Asthma Clin Immunol. 2006; 2:98–108.
Article40. Alves-Filho JC, Spiller F, Cunha FQ. Neutrophil paralysis in sepsis. Shock. 2010; 34:Suppl 1. 15–21.
Article41. Darcy CJ, Minigo G, Piera KA, Davis JS, McNeil YR, Chen Y, Volkheimer AD, Weinberg JB, Anstey NM, Woodberry T. Neutrophils with myeloid derived suppressor function deplete arginine and constrain T cell function in septic shock patients. Crit Care. 2014; 18:R163.
Article42. Janols H, Bergenfelz C, Allaoui R, Larsson AM, Rydén L, Björnsson S, Janciauskiene S, Wullt M, Bredberg A, Leandersson K. A high frequency of MDSCs in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J Leukoc Biol. 2014; 96:685–693.
Article43. Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol. 2016; 1:aaf8943.
Article44. Denny MF, Yalavarthi S, Zhao W, Thacker SG, Anderson M, Sandy AR, McCune WJ, Kaplan MJ. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol. 2010; 184:3284–3297.
Article45. Lood C, Blanco LP, Purmalek MM, Carmona-Rivera C, De Ravin SS, Smith CK, Malech HL, Ledbetter JA, Elkon KB, Kaplan MJ. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016; 22:146–153.
Article46. Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM, Rubin CJ, Zhao W, Olsen SH, Klinker M, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011; 187:538–552.
Article