Ann Clin Microbiol.
2017 Dec;20(4):90-96. 10.5145/ACM.2017.20.4.90.
Assessment of Efflux Activity Using H33342 Accumulation in Tigecycline-Resistant Acinetobacter baumannii Clinical Isolates
- Affiliations
-
- 1Premedical Science, College of Medicine, Chosun University, Gwangju, Korea.
- 2Department of Laboratory Medicine, College of Medicine, Chosun University, Gwangju, Korea. sjbjang@chosun.ac.kr
- 3Department of Internal Medicine, College of Medicine, Chosun University, Gwangju, Korea.
- 4Department of Urology, College of Medicine, Chosun University, Gwangju, Korea.
- 5Department of Medical Education, College of Medicine, Chosun University, Gwangju, Korea.
- KMID: 2398124
- DOI: http://doi.org/10.5145/ACM.2017.20.4.90
Abstract
- BACKGROUND
Tigecycline resistance has emerged recently and has shown diverse mechanisms. The aim of this study was to assess the role of efflux activity in tigecycline resistance in 120 clinical isolates of A. baumannii using two methods: the H33342 accumulation assay and adeB real-time reverse transcriptase polymerase chain reaction. In addition, we analyzed the correlation between the expression level of adeB and H33342 accumulation level.
METHODS
A. baumannii clinical isolates was divided into tigecycline-resistant (49 strains), intermediate (40 strains), and susceptible (31 strains) groups. The H33342 accumulation was measured in the absence or presence of the efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Real-time RT-PCR was performed to determine the relative expression of the adeB gene in A. baumannii clinical isolates.
RESULTS
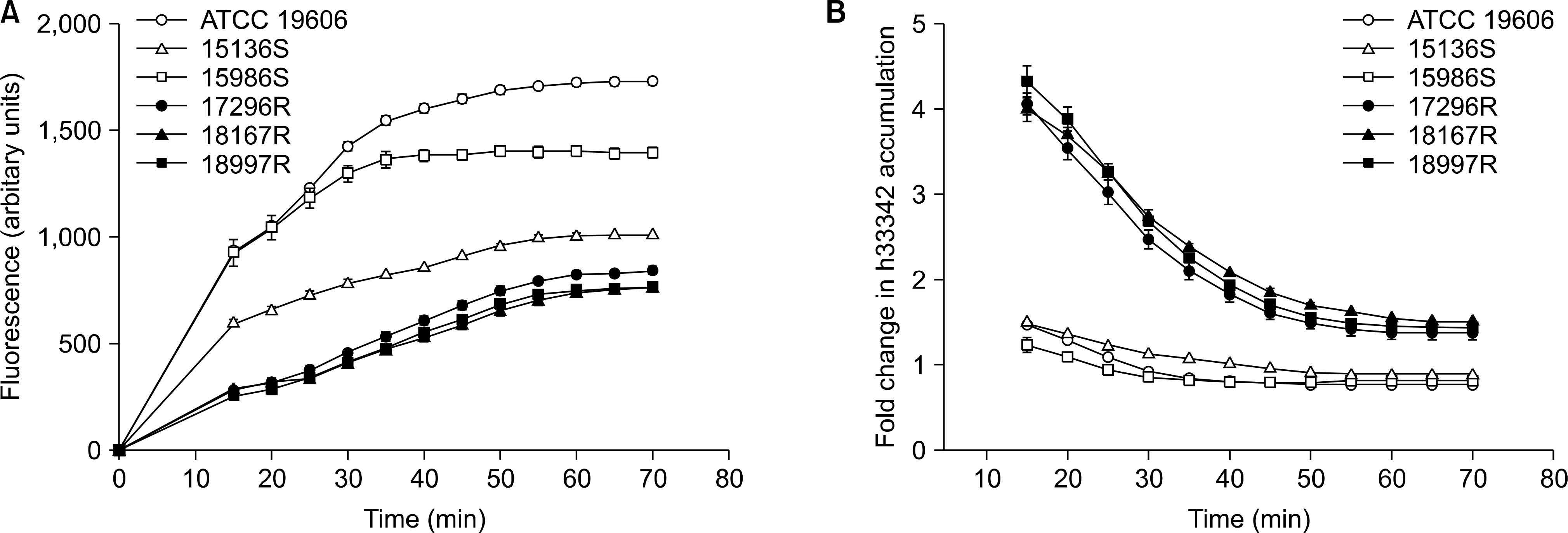
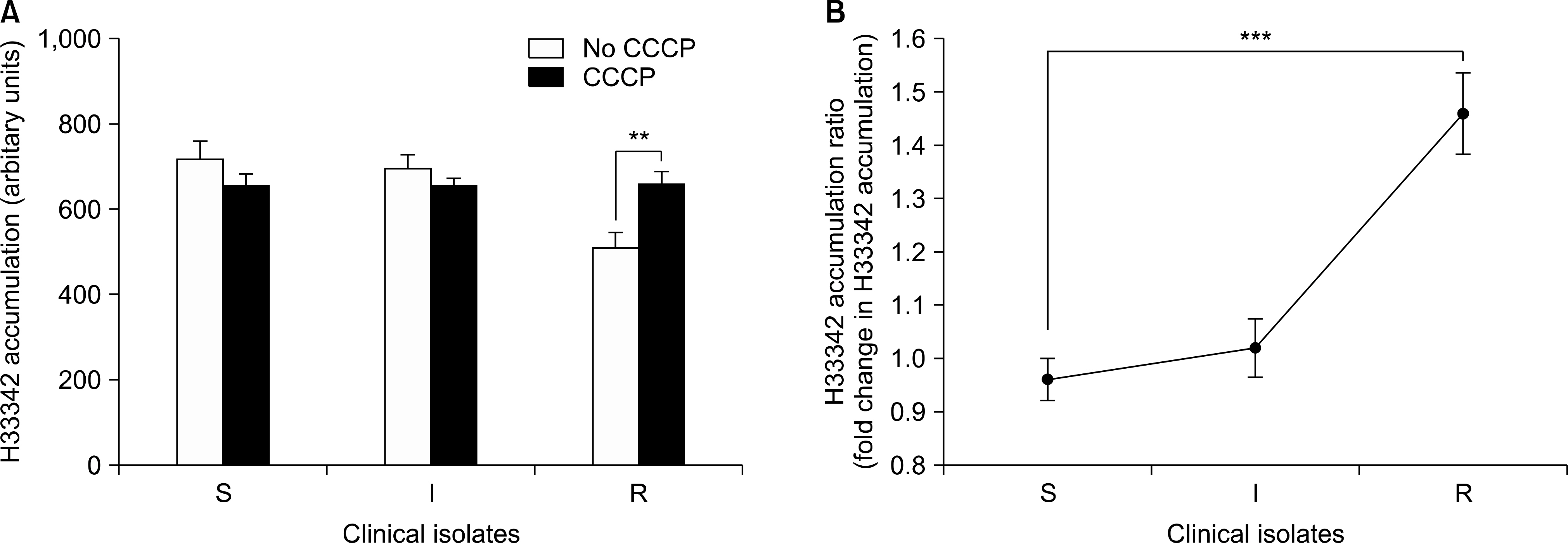
The level of H33342 accumulation in the resistant group was relatively lower than those in the other groups. The addition of CCCP caused a significantly increased fold change in H33342 accumulation in the tigecycline-resistant group. Significant difference in the fold change level in H33342 accumulation was found between tigecycline-susceptible and resistant isolates. Those findings support the role of efflux pumps of which substrates are H33342 in the resistance of tigecycline. Significant differences in the relative expression levels of adeB were shown between tigecycline-susceptible and resistant groups also.
CONCLUSION
The results showed that several efflux pumps of which substrates were H33342 can contribute to tigecycline resistance. The adeB overexpression can also contribute to tigecycline resistance. It is possible that efflux pumps other than adeB efflux pumps contribute to tigecycline resistance because there was no correlation between fold change level in H33342 accumulation and adeB expression level.
MeSH Terms
Figure
Reference
-
References
1. Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007; 5:939–51.
Article2. Coyne S, Courvalin P, Périchon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother. 2011; 55:947–53.3. Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, Dé E, et al. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio. 2015; 6:e00309–15.
Article4. Reid GE, Grim SA, Aldeza CA, Janda WM, Clark NM. Rapid development of Acinetobacter baumannii resistance to tigecycline. Pharmacotherapy. 2007; 27:1198–201.5. Deng M, Zhu MH, Li JJ, Bi S, Sheng ZK, Hu FS, et al. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother. 2014; 58:297–303.
Article6. Coldham NG, Webber M, Woodward MJ, Piddock LJ. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J Antimicrob Chemother. 2010; 65:1655–63.
Article7. Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D'Souza S, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014; 58:722–33.
Article8. Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001; 45:3375–80.9. Richmond GE, Chua KL, Piddock LJ. Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzamide). J Antimicrob Chemother. 2013; 68:1594–600.
Article10. Lee MJ, Jang SJ, Li XM, Park G, Kook JK, Kim MJ, et al. Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn Microbiol Infect Dis. 2014; 78:29–34.
Article11. Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of bla OXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008; 61:274–8.12. Kim HS, Kim DM, Neupane GP, Lee YM, Yang NW, Jang SJ, et al. Comparison of conventional, nested, and realtime PCR assays for rapid and accurate detection of Vibrio vulnificus. J Clin Microbiol. 2008; 46:2992–8.13. Whalen KE, Poulson-Ellestad KL, Deering RW, Rowley DC, Mincer TJ. Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3,4-dibromopyrrole-2,5-dione isolated from a Pseudoalteromonas sp. J Nat Prod. 2015; 78:402–12.
Article14. Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013; 57:5247–57.
Article15. Li XZ and Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009; 69:1555–623.16. Peleg AY, Adams J, Paterson DL. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007; 51:2065–9.17. Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013; 57:2989–95.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Extreme Drug Resistant Acinetobacter Nosocomial Ventilator-Associated Pneumonia Treated Successfully with Tigecycline and Amikacin in Intensive Care Unit: A Case Report
- Detection of Tigecycline Resistance in Acinetobacter baumannii: The Discrepancy between the Minimal Inhibitory Concentration Method and the Disk Diffusion Test
- Carbapenem-Resistant Acinetobacter baumannii
- In Vitro Interactions of Antibiotic Combinations of Colistin, Tigecycline, and Doripenem Against Extensively Drug-Resistant and Multidrug-Resistant Acinetobacter baumannii
- Correlation of Ciprofloxacin Resistance with the AdeABC Efflux System in Acinetobacter baumannii Clinical Isolates