Ann Lab Med.
2018 Jan;38(1):1-8. 10.3343/alm.2018.38.1.1.
Cell-Free DNA in Oncology: Gearing up for Clinic
- Affiliations
-
- 1Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA. CloudP_Paweletz@dfci.harvard.edu
- 2Belfer Center for Applied Cancer Science, Dana-Farber Cancer Institute, Boston, MA, USA.
- KMID: 2397595
- DOI: http://doi.org/10.3343/alm.2018.38.1.1
Abstract
- In the past several years, interest in the clinical utility of cell-free DNA as a noninvasive cancer biomarker has grown rapidly. Success in the development of plasma genotyping assays and other liquid biopsy assays has widened the scope of cell-free DNA use in research and the clinic. Already approved by the US Food and Drug Administration in the narrow context of epidermal growth factor receptor-mutated non-small cell lung cancer, plasma genotyping assays are currently being investigated in a wide array of clinical settings and modalities. These include plasma genotyping as a tool for early diagnosis, the detection of minimal residual disease, and the evaluation of treatment response/progression. In this review, we assess the clinical landscape of plasma genotyping assays and propose strategies for their further expansion into routine clinical care.
MeSH Terms
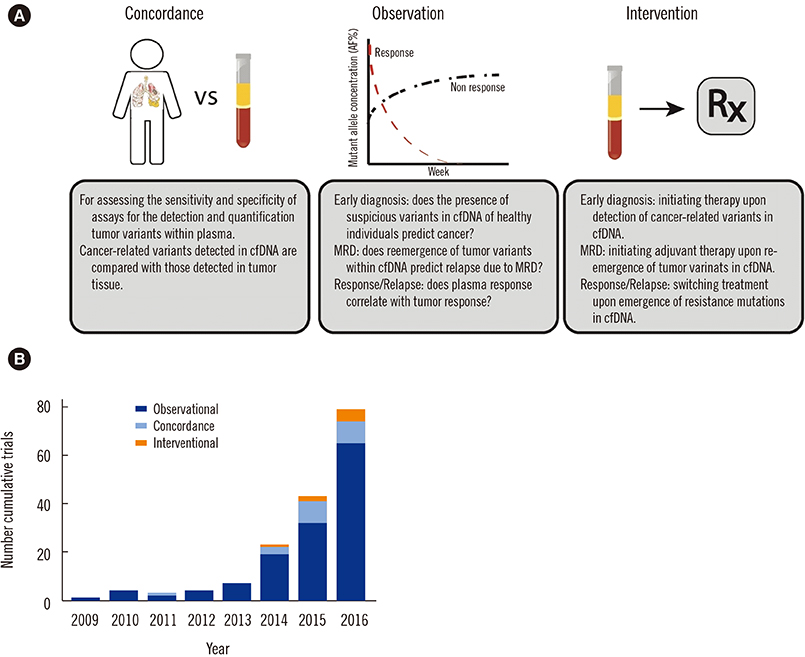
Figure
Cited by 2 articles
-
Targeted Next-Generation Sequencing of Plasma Cell-Free DNA in Korean Patients with Hepatocellular Carcinoma
Hyojin Chae, Pil Soo Sung, Hayoung Choi, Ahlm Kwon, Dain Kang, Yonggoo Kim, Myungshin Kim, Seung Kew Yoon
Ann Lab Med. 2021;41(2):198-206. doi: 10.3343/alm.2021.41.2.198.Application of Next Generation Sequencing in Laboratory Medicine
Yiming Zhong, Feng Xu, Jinhua Wu, Jeffrey Schubert, Marilyn M. Li
Ann Lab Med. 2021;41(1):25-43. doi: 10.3343/alm.2021.41.1.25.
Reference
-
1. Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez l'homme [in French]. C R Seances Soc Biol Fil. 1948; 142:241–243.2. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61:1659–1665.3. Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000; 906:161–168.
Article4. Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA. Clin Chim Acta. 2001; 313:139–142.
Article5. Papageorgiou EA, Karagrigoriou A, Tsaliki E, Velissariou V, Carter NP, Patsalis PC. Fetal-specific DNA methylation ratio permits noninvasive prenatal diagnosis of trisomy 21. Nat Med. 2011; 17:510–513.
Article6. Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350:485–487.
Article7. Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998; 339:1734–1738.
Article8. Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010; 2:20ra14.
Article9. Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012; 4:162ra154.
Article10. Center for Drug Evaluation and Research. Approved Drugs - cobas EGFR Mutation Test v2 [Internet]. U S Food and Drug Administration Home Page. Center for Drug Evaluation and Research;Updated on Jun 2016. https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm.11. Overman MJ, Modak J, Kopetz S, Murthy R, Yao JC, Hicks ME, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2013; 31:17–22.
Article12. Murtaza M, Dawson S-J, Tsui DWY, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013; 497:108–112.
Article13. Vanderlaan PA, Yamaguchi N, Folch E, Boucher DH, Kent MS, Gangadharan SP, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer. 2014; 84:39–44.
Article14. Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Ja Nne PA, Koch WH, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. 2016; 22:1103–1110.
Article15. Yanagita M, Redig AJ, Paweletz CP, Dahlberg SE, Oconnell A, Feeney N, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR-mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res. 2016; 22:6010–6020.
Article16. Oxnard GR, Paweletz CP, Kuang Y, Mach SL, Oconnell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014; 20:1698–1705.
Article17. Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014; 20:548–554.
Article18. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:9530–9535.
Article19. Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012; 109:14508–14513.
Article20. Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DWY, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012; 4:136ra68.
Article21. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999; 96:9236–9241.
Article22. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003; 100:8817–8822.
Article23. Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012; 18:1177–1185.
Article24. Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O'Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016; 2:1014–1022.25. The STRIVE Study: Breast Cancer Screening Cohort [Internet]. The STRIVE Study: Breast Cancer Screening Cohort - Full Text View -. ClinicalTrials.gov;Updated on Aug 2017. https://clinicaltrials.gov/ct2/show/NCT03085888?term=NCT03085888&rank=1.26. Utility of Plasma Circulating Tumor DNA (ctDNA) in Asymptomatic Subjects for the Detection of Neoplastic Disease (H1000) [Internet]. Utility of Plasma Circulating Tumor DNA (ctDNA) in Asymptomatic Subjects for the Detection of Neoplastic Disease - Full Text View -. ClinicalTrials;Updated on Apr 2017. https://clinicaltrials.gov/ct2/show/NCT02612350?term=NCT02612350&rank=1.27. Spanish Lung Liquid vs. Invasive Biopsy Program (SLLIP) [Internet]. Spanish Lung Liquid vs. Invasive Biopsy Program (SLLIP) - Full Text View -. ClinicalTrials.gov;Updated on Sep 2017. https://clinicaltrials.gov/ct2/show/NCT03248089?spons=Guardant&rank=1.28. Moyer VA. Screening for lung cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2014; 160:330–338.
Article29. U.S. Preventive Services Task Force. Screening for colorectal cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2008; 149:627.30. U.S. Preventive Services Task Force. Screening for breast cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2009; 151:716.31. Moyer VA. U.S. Preventive Services Task Force. Screening for cervical cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2012; 156:880.
Article32. Bettegowda C, Sausen M, Leary R, Kinde I, Agrawal N, Bartlett B, et al. Detection of circulating tumor DNA in early and late stage human malignancies. Sci Transl Med. 2014; 6:224ra24.
Article33. Science | GRAIL's Commitment to Scientific Rigor and Clinical Utility [Internet]. GRAIL;Update on 2017. https://grail.com/science/.34. Reporter S. Guardant Health, Pharma Firms to Develop 500-Gene Liquid Biopsy Panel [Internet]. GenomeWeb;2017. https://www.genomeweb.com/drug-discovery-development/guardant-health-pharma-firms-develop-500-gene-liquid-biopsy-panel.35. Gormally E. TP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective study. Cancer Res. 2006; 66:6871–6876.
Article36. Zhang BM, Aleshin A, Lin CY, Ford J, Zehnder JL, Suarez CJ. IDH2 mutation in a patient with metastatic colon cancer. N Engl J Med. 2017; 376:1991–1992.
Article37. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature. 2017; 545:446–451.38. Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016; 113:E1826–E1834.
Article39. Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for non-invasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015; 112:E5503–E5512.
Article40. Kang S, Li Q, Chen Q, Zhou Y, Park S, Lee G, et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 2017; 18:53.
Article41. Alliance for Clinical Trials in Oncology [Internet]. Alliance Trials – Full Text View --. https://www.allianceforclinicaltrialsinoncology.org. https://www.allianceforclinicaltrialsinoncology.org/main/public/standard.xhtml?path=%2FPublic%2FAlliance-Trials.42. Zimmermann FB, Molls M, Jeremic B. Treatment of recurrent disease in lung cancer. Semin Surg Oncol. 2003; 21:122–127.
Article43. Sadahiro S, Suzuki T, Ishikawa K, Nakamura T, Tanaka Y, Masuda T, et al. Recurrence patterns after curative resection of colorectal cancer in patients followed for a minimum of ten years. Hepatogastroenterology. 2003; 50:1362–1366.44. Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013; 10:460–471.
Article45. Figueredo A, Rumble RB, Maroun J, Earle CC, Cummings B, Mcleod R, et al. Follow-up of patients with curatively resected colorectal cancer: a practice guideline. BMC Cancer. 2003; 3:26.
Article46. Pita-Fernandez S, Alhayek-Ai M, Gonzalez-Martin C, Lopez-Calvino B, Seoane-Pillado T, Pertega-Diaz S. Intensive follow-up strategies improve outcomes in nonmetastatic colorectal cancer patients after curative surgery: a systematic review and meta-analysis. Ann Oncol. 2014; 26:644–656.47. Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. J Clin Oncol. 2009; 27:e279–e280.
Article48. Benson AB 3rd, Desch CE, Flynn PJ, Krause C, Loprinzi CL, Minsky BD, et al. 2000 Update of American Society of Clinical Oncology Colorectal Cancer Surveillance Guidelines. J Clin Oncol. 2000; 18:3586–3588.
Article49. Olsson E, Winter C, George A, Chen Y, Howlin J, Tang M-HE, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015; 7:1034–1047.50. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015; 7:302ra133.
Article51. Beaver JA, Jelovac D, Balukrishna S, Cochran RL, Croessmann S, Zabransky DJ, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014; 20:2643–2650.
Article52. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016; 8:346ra92.
Article53. Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2015; 65:625–634.
Article54. Frattini M, Gallino G, Signoroni S, Balestra D, Lusa L, Battaglia L, et al. Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett. 2008; 263:170–181.
Article55. Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015; 16:541–549.
Article56. Kim K, Shin DG, Park MK, Baik SH, Kim TH, Kim S, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res. 2014; 86:136.
Article57. Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015; 6:7686.
Article58. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2007; 14:985–990.
Article59. Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012; 486:537–540.
Article60. Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non–small-cell lung cancer. J Clin Oncol. 2016; 34:3375–3382.
Article61. Blood Sample Monitoring of Patients With EGFR Mutated Lung Cancer [Internet]. Blood Sample Monitoring of Patients with EGFR Mutated Lung Cancer - Full Text View -. ClinicalTrials.gov;Update on Mar 2017. https://clinicaltrials.gov/ct2/show/NCT02284633?term=NCT02284633&rank=1.62. Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015; 7:131ra182.
Article63. Lallous N, Volik SV, Awrey S, Leblanc E, Tse R, Murillo J, et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 2016; 17:10.
Article64. Azad AA, Volik SV, Wyatt AW, Haegert A, Bihan SL, Bell RH, et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res. 2015; 21:2315–2324.
Article65. Chu D, Paoletti C, Gersch C, Vandenberg DA, Zabransky DJ, Cochran RL, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res. 2016; 22:993–999.
Article66. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015; 21:795–801.
Article67. Morelli MP, Overman MJ, Dasari A, Kazmi SMA, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015; 26:731–736.
Article68. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012; 486:532–536.
Article69. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19:2240–2247.
Article70. Press Announcements - FDA approves new pill to treat certain patients with non-small cell lung cancer [Internet]. U S Food and Drug Administration Home Page. Updated on Nov 2015. https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm472525.htm.71. Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015; 372:1689–1699.
Article72. Sequist LV, Soria JC, Goldman JW, Wakelee HA, Gadgeel SM, Varga A, et al. Rociletinib in EGFR-mutated non–small-cell lung cancer. N Engl J Med. 2015; 372:1700–1709.
Article73. Guibert N, Pradines A, Farella M, Casanova A, Gouin S, Keller L, et al. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS-mutated lung adenocarcinoma. Lung Cancer. 2016; 100:1–4.74. Guibert N, Pradines A, Casanova A, Farella M, Keller L, Soria JC, et al. Detection and monitoring of the BRAF mutation in circulating tumor cells and circulating tumor DNA in BRAF-mutated lung adenocarcinoma. J Thorac Oncol. 2016; 11:e109–e112.75. Gray ES, Rizos H, Reid AL, Boyd SC, Pereira MR, Lo J, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015; 6:42008–42018.
Article76. Bidard FC, Madic J, Mariani P, Piperno-Neumann S, Rampanou A, Servois V, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2013; 134:1207–1213.
Article77. Paweletz CP, Oxnard GR, Feeney N, Hilton JF, Gandhi L, Do KT, et al. Serial droplet digital PCR (ddPCR) of plasma cell-free DNA (cfDNA) as pharmacodynamic (PD) biomarker in Phase 1 clinical trials for patients (pts) with KRAS mutant non-small cell lung cancer (NSCLC). In : American Association for Cancer Research Annual Meeting in 2016;
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Mitochondria and Aging
- Non-invasive prenatal test using cell free DNA
- Advantages of the single nucleotide polymorphism-based noninvasive prenatal test
- Clinical Circulating Tumor DNA Testing for Precision Oncology
- Next-Generation Sequencing-Based Molecular Profiling Using Cell-Free DNA: A Valuable Tool for the Diagnostic and Prognostic Evaluation of Patients With Gastric Cancer