Cancer Res Treat.
2023 Apr;55(2):351-366. 10.4143/crt.2022.1026.
Clinical Circulating Tumor DNA Testing for Precision Oncology
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- KMID: 2541224
- DOI: http://doi.org/10.4143/crt.2022.1026
Abstract
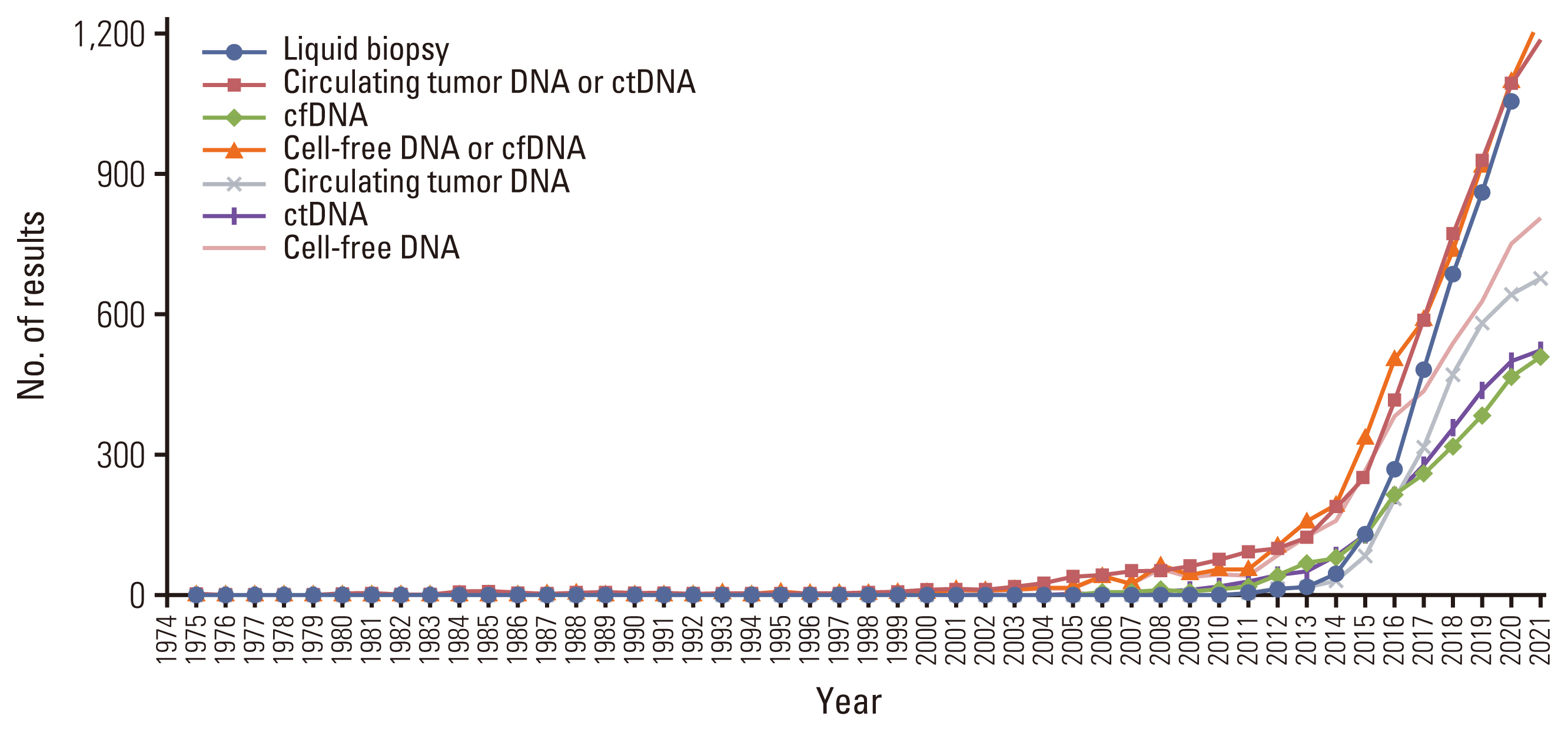
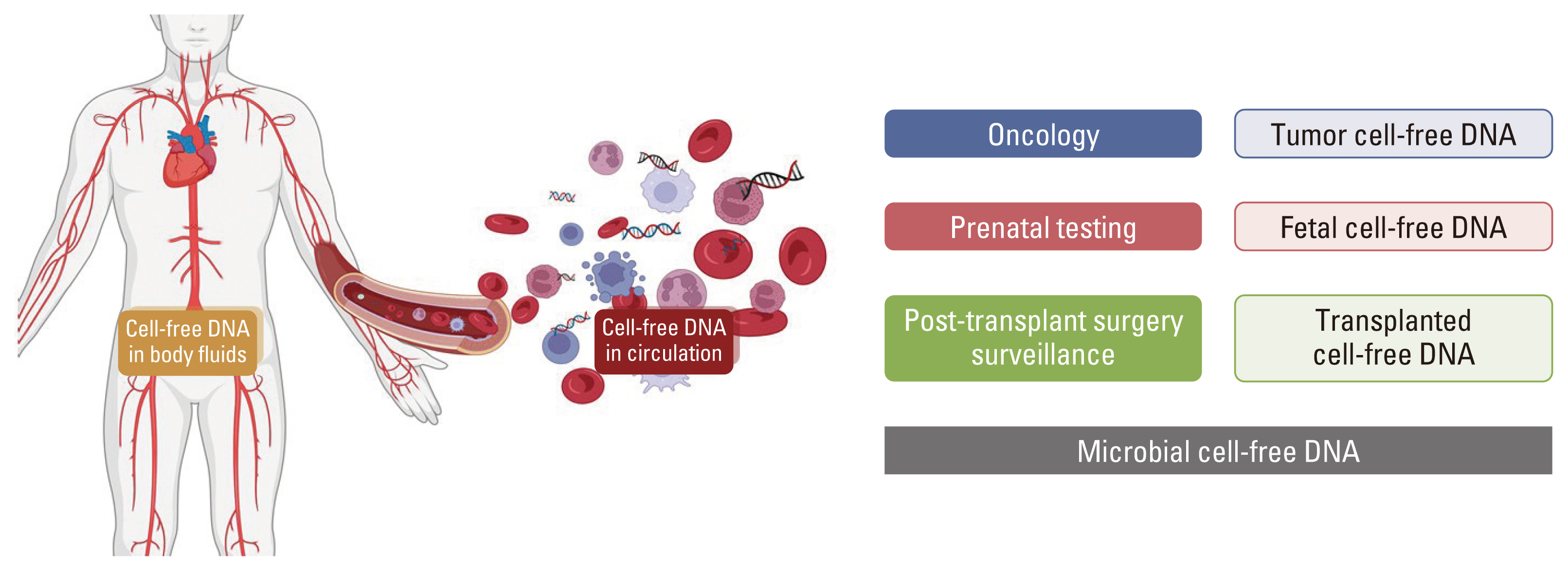
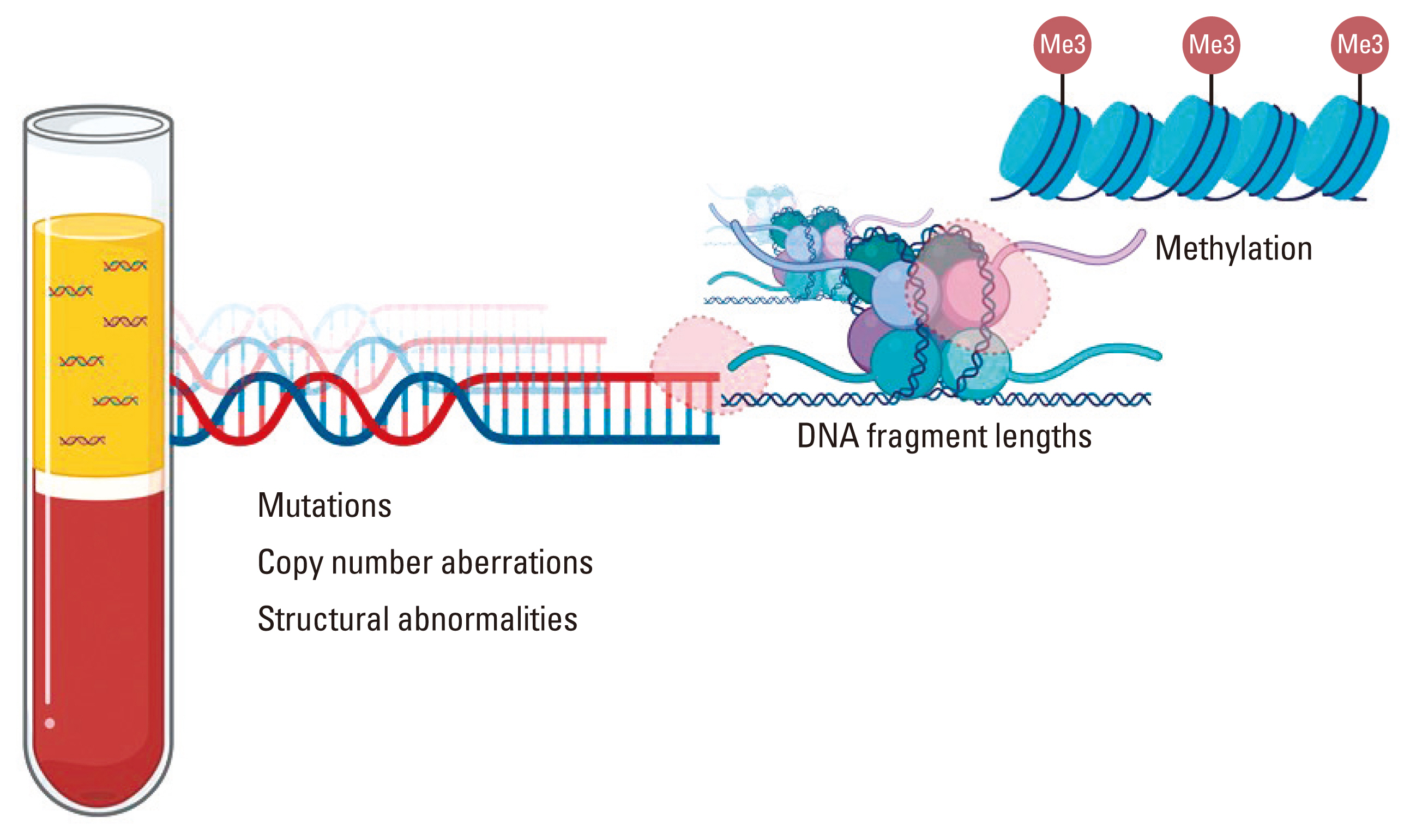
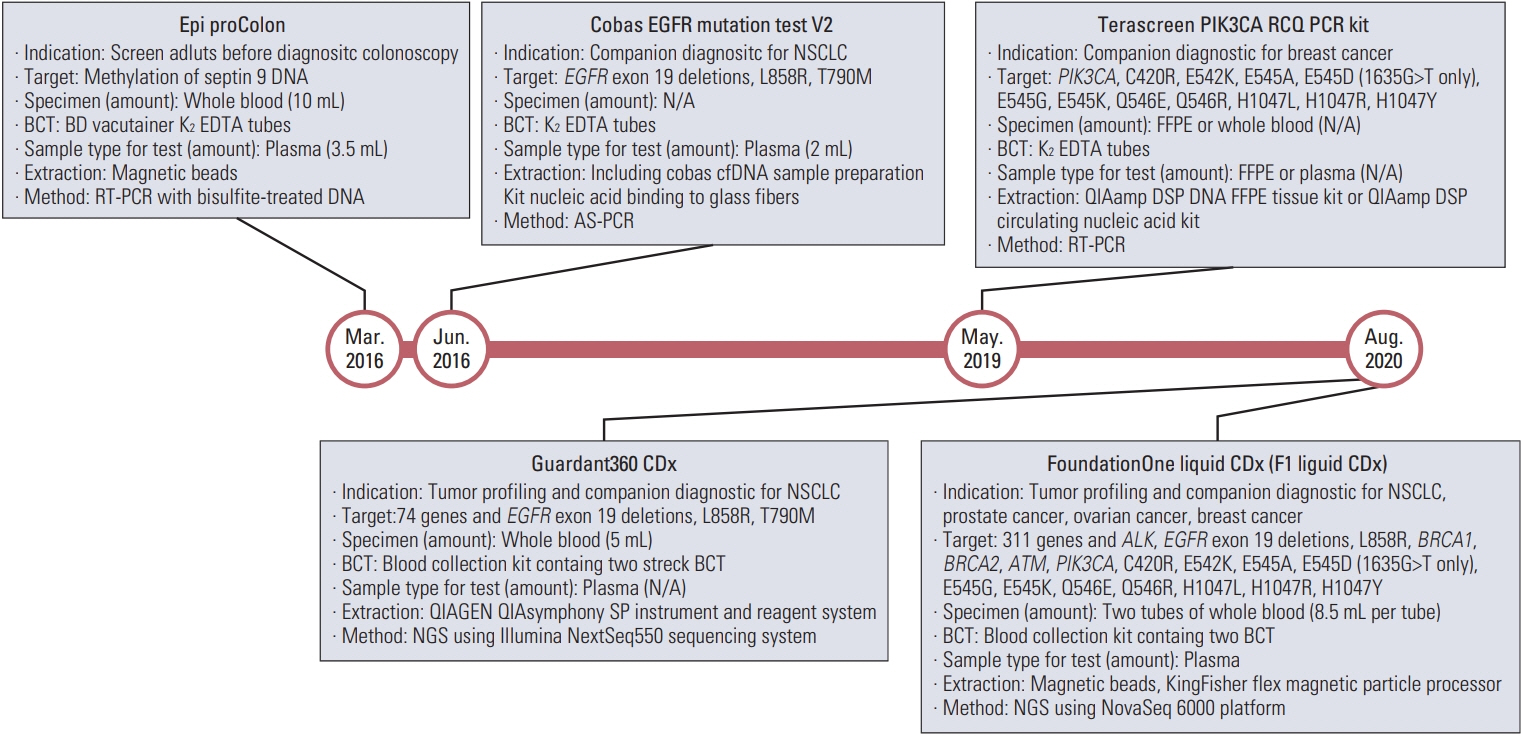
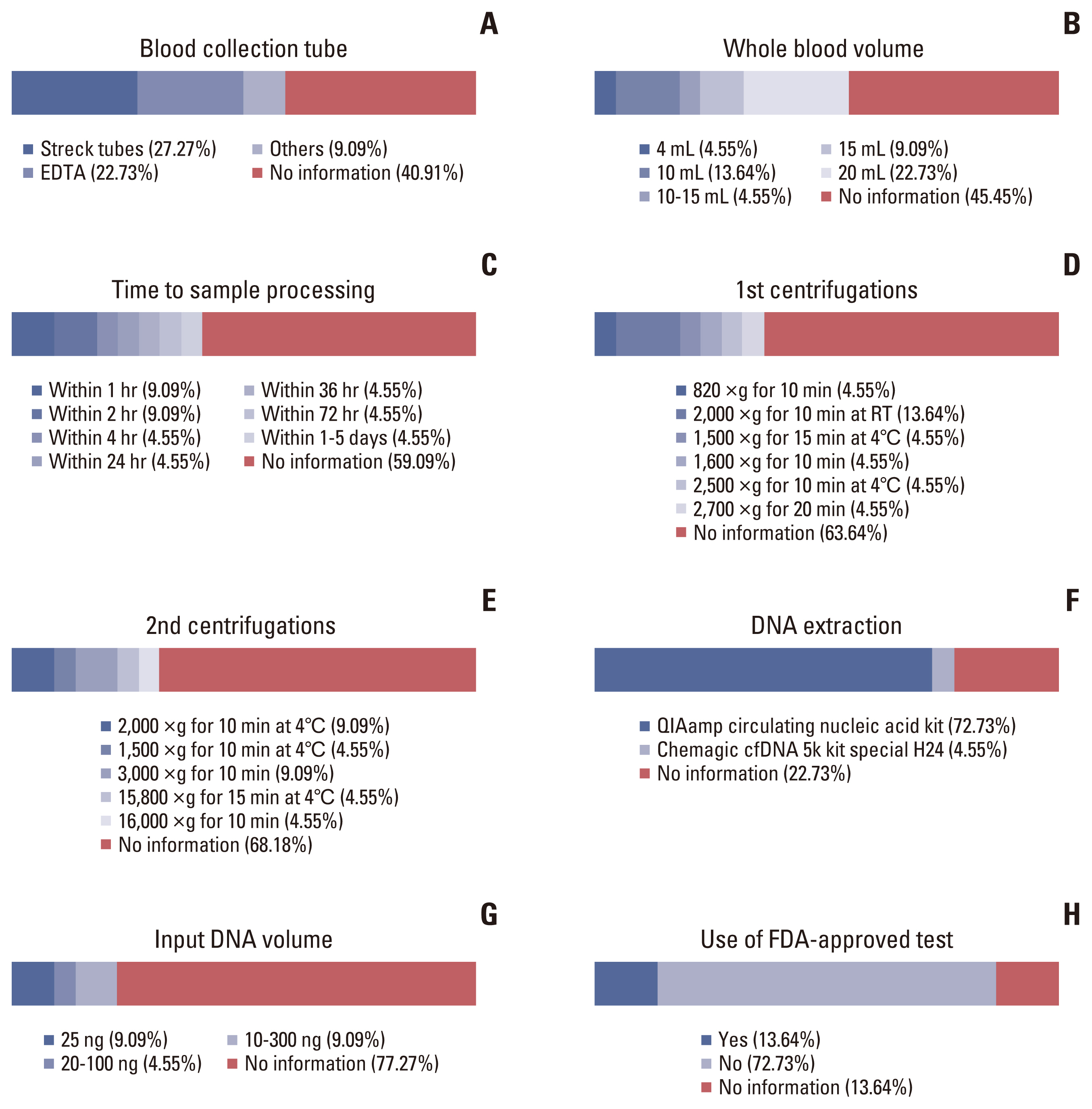
- Circulating tumor DNA (ctDNA) is the portion of the cell-free DNA in the blood of cancer patients released from tumor cells via apoptosis, necrosis, or active release. From 10 mL of blood, the 4-5 mL of plasma obtained from a cancer patient contains 5-10 ng/mL of ctDNA. The plasma contains not only ctDNA of tumor origin, but also DNA from normal cells or clonal hematopoiesis. Another characteristic of ctDNA is its rapid clearance from circulation; it has a half-life of 16 minutes to 2.5 hours. Obtaining reliable results from ctDNA requires the application and approval of standardized clinical validation guidelines; however, the status of numerous ctDNA tests currently varies. The clinical use of ctDNA testing should be carefully considered based on the test’s specific needs and characteristics. Here we provide the different characteristics of ctDNA tests and information regarding their validation and approval status.
Figure
Cited by 1 articles
-
Clinical Practice Recommendations for the Use of Next-Generation Sequencing in Patients with Solid Cancer: A Joint Report from KSMO and KSP
Miso Kim, Hyo Sup Shim, Sheehyun Kim, In Hee Lee, Jihun Kim, Shinkyo Yoon, Hyung-Don Kim, Inkeun Park, Jae Ho Jeong, Changhoon Yoo, Jaekyung Cheon, In-Ho Kim, Jieun Lee, Sook Hee Hong, Sehhoon Park, Hyun Ae Jung, Jin Won Kim, Han Jo Kim, Yongjun Cha, Sun Min Lim, Han Sang Kim, Choong-kun Lee, Jee Hung Kim, Sang Hoon Chun, Jina Yun, So Yeon Park, Hye Seung Lee, Yong Mee Cho, Soo Jeong Nam, Kiyong Na, Sun Och Yoon, Ahwon Lee, Kee-Taek Jang, Hongseok Yun, Sungyoung Lee, Jee Hyun Kim, Wan-Seop Kim
Cancer Res Treat. 2024;56(3):721-742. doi: 10.4143/crt.2023.1043.
Reference
-
References
1. Mandel P, Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948; 142:241–3.2. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994; 3:67–71.3. Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996; 2:1035–7.
Article4. Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001; 313:139–42.5. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014; 32:579–86.
Article6. Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010; 16:398–406.
Article7. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005; 102:16368–73.
Article8. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61:1659–65.9. Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016; 2:1014–22.
Article10. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008; 14:985–90.
Article11. Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006; 24:4270–6.
Article12. Li L, Hann HW, Wan S, Hann RS, Wang C, Lai Y, et al. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci Rep. 2016; 6:23992.
Article13. Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016; 113:E1826–34.
Article14. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–37.
Article15. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014; 20:1472–8.
Article16. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014; 371:2477–87.
Article17. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014; 371:2488–98.
Article18. Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.
Article19. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014; 6:224ra24.20. Cisneros-Villanueva M, Hidalgo-Perez L, Rios-Romero M, Cedro-Tanda A, Ruiz-Villavicencio CA, Page K, et al. Cell-free DNA analysis in current cancer clinical trials: a review. Br J Cancer. 2022; 126:391–400.
Article21. Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013; 73:6384–8.
Article22. Bronkhorst AJ, Ungerer V, Diehl F, Anker P, Dor Y, Fleischhacker M, et al. Towards systematic nomenclature for cell-free DNA. Hum Genet. 2021; 140:565–78.
Article23. BioRender. Create professional science figures in minutes [Internet]. Toronto, ON: BioRender;2022. [cited 2022 Oct 10]. Available from: https://biorender.com/.24. Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021; 124:345–58.
Article25. Gray ES, Witkowski T, Pereira M, Calapre L, Herron K, Irwin D, et al. Genomic analysis of circulating tumor DNA using a melanoma-specific UltraSEEK Oncogene Panel. J Mol Diagn. 2019; 21:418–26.26. Kohn L, Johansson M, Grankvist K, Nilsson J. Liquid biopsies in lung cancer-time to implement research technologies in routine care? Ann Transl Med. 2017; 5:278.
Article27. Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006; 3:551–9.
Article28. Sorber L, Zwaenepoel K, Deschoolmeester V, Van Schil PE, Van Meerbeeck J, Lardon F, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer. 2017; 107:100–7.
Article29. Santis G, Angell R, Nickless G, Quinn A, Herbert A, Cane P, et al. Screening for EGFR and KRAS mutations in endobronchial ultrasound derived transbronchial needle aspirates in non-small cell lung cancer using COLD-PCR. PLoS One. 2011; 6:e25191.
Article30. Castellanos-Rizaldos E, Liu P, Milbury CA, Guha M, Brisci A, Cremonesi L, et al. Temperature-tolerant COLD-PCR reduces temperature stringency and enables robust mutation enrichment. Clin Chem. 2012; 58:1130–8.
Article31. Narayan A, Carriero NJ, Gettinger SN, Kluytenaar J, Kozak KR, Yock TI, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012; 72:3492–8.
Article32. Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016; 34:547–55.
Article33. Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003; 100:8817–22.
Article34. Dang DK, Park BH. Circulating tumor DNA: current challenges for clinical utility. J Clin Invest. 2022; 132:e154941.
Article35. Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019; 13:34.
Article36. Mosko MJ, Nakorchevsky AA, Flores E, Metzler H, Ehrich M, van den Boom DJ, et al. Ultrasensitive detection of multiplexed somatic mutations using MALDI-TOF mass spectrometry. J Mol Diagn. 2016; 18:23–31.
Article37. Arisi MF, Dotan E, Fernandez SV. Circulating tumor DNA in precision oncology and its applications in colorectal cancer. Int J Mol Sci. 2022; 23:4441.
Article38. Baer C, Kern W, Koch S, Nadarajah N, Schindela S, Meggendorfer M, et al. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica. 2016; 101:830–8.
Article39. Soda N, Clack K, Shiddiky MJ. Recent advances in liquid biopsy technologies for cancer biomarker detection. Sens Diagn. 2022; 1:343–75.
Article40. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016; 12:e1006162.
Article41. Chen A, Li J, Wang L, Huang Q, Zhu J, Wen S, et al. Comparison of paired cerebrospinal fluid and serum cell-free mitochondrial and nuclear DNA with copy number and fragment length. J Clin Lab Anal. 2020; 34:e23238.
Article42. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019; 570:385–9.
Article43. Cai Z, Wang Z, Liu C, Shi D, Li D, Zheng M, et al. Detection of microsatellite instability from circulating tumor DNA by targeted deep sequencing. J Mol Diagn. 2020; 22:860–70.44. Arzimanoglou II, Gilbert F, Barber HR. Microsatellite instability in human solid tumors. Cancer. 1998; 82:1808–20.
Article45. Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017; 8:15180.
Article46. Boyiadzis MM, Kirkwood JM, Marshall JL, Pritchard CC, Azad NS, Gulley JL. Significance and implications of FDA approval of pembrolizumab for biomarker-defined disease. J Immunother Cancer. 2018; 6:35.
Article47. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014; 30:1015–6.
Article48. Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014; 60:1192–9.
Article49. Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017; 8:7452–63.
Article50. Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013; 339:1567–70.
Article51. Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020; 12:eaax7533.
Article52. Ofman JJ, Hall MP, Aravanis AM. GRAIL and the quest for earlier multi-cancer detection [Internet]. Berlin: Springer Nature;2018. [cited 2022 Oct 10]. Available from: https://www.nature.com/articles/d42473-020-00079-y.53. Koch A, Joosten SC, Feng Z, de Ruijter TC, Draht MX, Melotte V, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018; 15:459–66.
Article54. Daniunaite K, Jarmalaite S, Kriukiene E. Epigenomic technologies for deciphering circulating tumor DNA. Curr Opin Biotechnol. 2019; 55:23–9.
Article55. Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003; 35:146–50.
Article56. Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003; 35:152–6.
Article57. Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999; 59:71–3.58. Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008; 3:1903–8.
Article59. Xia Y, Tang W, Qian X, Li X, Cheng F, Wang K, et al. Efficacy and safety of camrelizumab plus apatinib during the perioperative period in resectable hepatocellular carcinoma: a single-arm, open label, phase II clinical trial. J Immunother Cancer. 2022; 10:e004656.
Article60. Openshaw MR, Mohamed AA, Ottolini B, Fernandez-Garcia D, Richards CJ, Page K, et al. Longitudinal monitoring of circulating tumour DNA improves prognostication and relapse detection in gastroesophageal adenocarcinoma. Br J Cancer. 2020; 123:1271–9.
Article61. Martinez-Saez O, Pascual T, Braso-Maristany F, Chic N, Gonzalez-Farre B, Sanfeliu E, et al. Circulating tumor DNA dynamics in advanced breast cancer treated with CDK4/6 inhibition and endocrine therapy. NPJ Breast Cancer. 2021; 7:8.
Article62. Engel T, Ben-Horin S, Beer-Gabel M. Autonomic dysfunction correlates with clinical and inflammatory activity in patients with Crohn’s disease. Inflamm Bowel Dis. 2015; 21:2320–6.
Article63. Wang X, Liu H, Zhao C, Li W, Xu H, Chen Y. The DEAD-box RNA helicase 51 controls non-small cell lung cancer proliferation by regulating cell cycle progression via multiple pathways. Sci Rep. 2016; 6:26108.
Article64. Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. Arch Pathol Lab Med. 2018; 142:1242–53.
Article65. El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta. 2013; 424:222–30.
Article66. Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013; 46:1561–5.
Article67. Toro PV, Erlanger B, Beaver JA, Cochran RL, VanDenBerg DA, Yakim E, et al. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin Biochem. 2015; 48:993–8.
Article68. Barra GB, Santa Rita TH, de Almeida Vasques J, Chianca CF, Nery LF, Santana Soares Costa S. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin Biochem. 2015; 48:976–81.
Article69. Li D, Kusko R, Ning B, Tong W, Johann DJ Jr, Xu J. FDA-led consortium studies advance quality control of targeted next generation sequencing assays for precision oncology. Precis Cancer Med. 2021; 4:32.
Article70. Gong B, Li D, Kusko R, Novoradovskaya N, Zhang Y, Wang S, et al. Cross-oncopanel study reveals high sensitivity and accuracy with overall analytical performance depending on genomic regions. Genome Biol. 2021; 22:109.71. Deveson IW, Gong B, Lai K, LoCoco JS, Richmond TA, Schageman J, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. 2021; 39:1115–28.
Article72. Zhang Y, Blomquist TM, Kusko R, Stetson D, Zhang Z, Yin L, et al. Deep oncopanel sequencing reveals within block position-dependent quality degradation in FFPE processed samples. Genome Biol. 2022; 23:141.
Article73. Willey JC, Morrison TB, Austermiller B, Crawford EL, Craig DJ, Blomquist TM, et al. Advancing NGS quality control to enable measurement of actionable mutations in circulating tumor DNA. Cell Rep Methods. 2021; 1:100106.
Article74. U.S. Food Drug Administration. Drug Approvals and database [Internet]. Silver Spring MD: U.S. Food and Drug Administration;2020. [cited 2020 Oct 10]. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers.75. Duffy MJ, Crown J. Use of circulating tumour DNA (ctDNA) for measurement of therapy predictive biomarkers in patients with cancer. J Pers Med. 2022; 12:99.
Article76. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. NCCN guidelines insights: non-small cell lung cancer, version 2.2021. J Natl Compr Canc Netw. 2021; 19:254–66.77. Corcoran RB. Liquid biopsy versus tumor biopsy for clinical-trial recruitment. Nat Med. 2020; 26:1815–6.
Article78. Ghosh RK, Pandey T, Dey P. Liquid biopsy: a new avenue in pathology. Cytopathology. 2019; 30:138–43.
Article79. Marrugo-Ramirez J, Mir M, Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int J Mol Sci. 2018; 19:2877.
Article80. Chae YK, Oh MS. Detection of minimal residual disease using ctDNA in lung cancer: current evidence and future directions. J Thorac Oncol. 2019; 14:16–24.
Article81. Moding EJ, Nabet BY, Alizadeh AA, Diehn M. Detecting liquid remnants of solid tumors: circulating tumor DNA minimal residual disease. Cancer Discov. 2021; 11:2968–86.
Article82. Pellini B, Chaudhuri AA. Circulating tumor DNA minimal residual disease detection of non-small-cell lung cancer treated with curative intent. J Clin Oncol. 2022; 40:567–75.
Article83. Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res. 2021; 27:5586–94.84. Menikoff J, Kaneshiro J, Pritchard I. The common rule, updated. N Engl J Med. 2017; 376:613–5.
Article85. Dresser R. Research ethics. Aligning regulations and ethics in human research. Science. 2012; 337:527–8.86. Detsky AS. Sources of bias for authors of clinical practice guidelines. CMAJ. 2006; 175:1033–5.
Article87. Sturgeon CM, Diamandis E. Laboratory medicine practice guidelines. Use of tumor markers in clinical practice: quality requirements. Washington, DC: National Academy of Clinical Biochemistry;2009.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Liquid Biopsy: Current Status and Future Perspective in Gastric Cancer and Helicobacter Infection
- Exploring the prognostic value of ultra-low-pass whole-genome sequencing of circulating tumor DNA in hepatocellular carcinoma
- Utilizing Plasma Circulating Tumor DNA Sequencing for Precision Medicine in the Management of Solid Cancers
- Circulating Cell-free Tumor Nucleic Acids in Gastric Cancer