Molecular Testing of Lung Cancers
- Affiliations
-
- 1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea.
- 2Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Department of Pathology, Inha University School of Medicine, Incheon, Korea.
- 4Department of Pathology, Inje University Ilsan Paik Hospital, Inje University, Goyang, Korea.
- 5Department of Pathology, Konkuk University School of Medicine, Seoul, Korea.
- 6Department of Pathology, Dong-A University College of Medicine, Busan, Korea.
- 7Department of Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 8Department of Pathology, Gachon University Gil Medical Center, Incheon, Korea.
- 9Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea.
- 10Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- 11Department of Pathology, National Cancer Center, Goyang, Korea. gklee@ncc.re.kr
- KMID: 2392592
- DOI: http://doi.org/10.4132/jptm.2017.04.10
Abstract
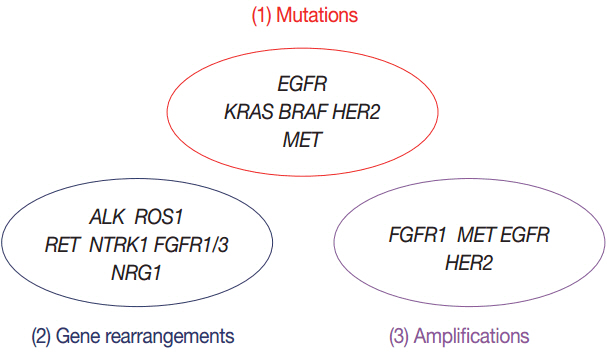
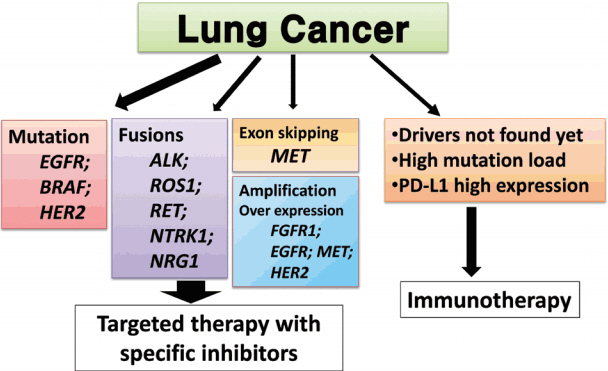
- Targeted therapies guided by molecular diagnostics have become a standard treatment of lung cancer. Epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements are currently used as the best predictive biomarkers for EGFR tyrosine kinase inhibitors and ALK inhibitors, respectively. Besides EGFR and ALK, the list of druggable genetic alterations has been growing, including ROS1 rearrangements, RET rearrangements, and MET alterations. In this situation, pathologists should carefully manage clinical samples for molecular testing and should do their best to quickly and accurately identify patients who will benefit from precision therapeutics. Here, we grouped molecular biomarkers of lung cancers into three categories"”mutations, gene rearrangements, and amplifications"”and propose expanded guidelines on molecular testing of lung cancers.
MeSH Terms
Figure
Cited by 5 articles
-
Human Leukocyte Antigen Class I and Programmed Death-Ligand 1 Coexpression Is an Independent Poor Prognostic Factor in Adenocarcinoma of the Lung
Yeon Bi Han, Hyun Jung Kwon, Soo Young Park, Eun-Sun Kim, Hyojin Kim, Jin-Haeng Chung
J Pathol Transl Med. 2019;53(2):86-93. doi: 10.4132/jptm.2018.12.26.Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
J Pathol Transl Med. 2021;55(3):181-191. doi: 10.4132/jptm.2021.03.23.Detection of Targetable Genetic Alterations in Korean Lung Cancer Patients: A Comparison Study of Single-Gene Assays and Targeted Next-Generation Sequencing
Eunhyang Park, Hyo Sup Shim
Cancer Res Treat. 2020;52(2):543-551. doi: 10.4143/crt.2019.305.TM4SF4 and LRRK2 Are Potential Therapeutic Targets in Lung and Breast Cancers through Outlier Analysis
Kyungsoo Jung, Joon-Seok Choi, Beom-Mo Koo, Yu Jin Kim, Ji-Young Song, Minjung Sung, Eun Sol Chang, Ka-Won Noh, Sungbin An, Mi-Sook Lee, Kyoung Song, Hannah Lee, Ryong Nam Kim, Young Kee Shin, Doo-Yi Oh, Yoon-La Choi
Cancer Res Treat. 2021;53(1):9-24. doi: 10.4143/crt.2020.434.Landscape of
EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
J Pathol Transl Med. 2022;56(5):249-259. doi: 10.4132/jptm.2022.06.11.
Reference
-
1. Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013; 31:1039–49.
Article2. Shim HS, Chung JH, Kim L, et al. Guideline recommendations for EGFR mutation testing in lung cancer: proposal of the Korean Cardiopulmonary Pathology Study Group. Korean J Pathol. 2013; 47:100–6.3. Kim H, Shim HS, Kim L, et al. Guideline recommendations for testing of ALK gene rearrangement in lung cancer: a proposal of the Korean Cardiopulmonary Pathology Study Group. Korean J Pathol. 2014; 48:1–9.4. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–25.5. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–50.6. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015; 524:47–53.7. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009; 361:947–57.
Article8. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010; 362:2380–8.9. Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015; 10:438–45.10. Lee SH, Kim WS, Choi YD, et al. Analysis of mutations in epidermal growth factor receptor gene in Korean patients with non-small vell lung cancer: summary of a nationwide survey. J Pathol Transl Med. 2015; 49:481–8.11. Lee B, Lee T, Lee SH, Choi YL, Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016; 7:23874–84.12. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011; 3:75ra26.
Article13. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013; 19:2240–7.14. Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014; 11:473–81.
Article15. Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res. 2011; 17:1169–80.
Article16. Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015; 372:1689–99.
Article17. Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015; 21:560–2.18. Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol. 2015; 26:2073–8.19. Lee B, Lee B, Han G, Kwon MJ, Han J, Choi YL. KRAS mutation detection in non-small cell lung cancer using a peptide nucleic acid-mediated polymerase chain reaction clamping method and comparative validation with next-generation sequencing. Korean J Pathol. 2014; 48:100–7.20. Kim HR, Ahn JR, Lee JG, et al. The impact of cigarette smoking on the frequency of and qualitative differences in KRAS mutations in Korean patients with lung adenocarcinoma. Yonsei Med J. 2013; 54:865–74.21. Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016; 2:805–12.22. Shim HS, Kenudson M, Zheng Z, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol. 2015; 10:1156–62.
Article23. Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011; 29:3574–9.24. Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011; 29:2046–51.25. Ha SY, Choi SJ, Cho JH, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget. 2015; 6:5465–74.
Article26. Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 2016; 17:984–93.27. Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016; 17:642–50.28. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015; 373:726–36.29. Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013; 31:1997–2003.30. Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015; 5:842–9.31. Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol. 2016; 34:721–30.32. D’Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011; 29:2066–70.33. Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012; 7:323–30.34. Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol. 2010; 5:1706–13.35. Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011; 6:244–85.36. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013; 8:823–59.
Article37. Noh S, Shim H. Optimal combination of immunohistochemical markers for subclassification of non-small cell lung carcinomas: a tissue microarray study of poorly differentiated areas. Lung Cancer. 2012; 76:51–5.
Article38. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007; 13:4954–5.39. Lee HJ, Xu X, Kim H, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol. 2013; 47:52–60.40. Kim HJ, Lee KY, Kim YC, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer. 2012; 75:321–5.
Article41. Han HS, Lim SN, An JY, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012; 7:355–64.42. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015; 5:850–9.
Article43. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013; 31:1023–31.
Article44. Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014; 20:1479–84.
Article45. Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011; 6:451–8.
Article46. Navani N, Brown JM, Nankivell M, et al. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med. 2012; 185:1316–22.47. Sun PL, Jin Y, Kim H, Lee CT, Jheon S, Chung JH. High concordance of EGFR mutation status between histologic and corresponding cytologic specimens of lung adenocarcinomas. Cancer Cytopathol. 2013; 121:311–9.48. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011; 29:2972–7.49. Chung JH, Choe G, Jheon S, et al. Epidermal growth factor receptor mutation and pathologic-radiologic correlation between multiple lung nodules with ground-glass opacity differentiates multicentric origin from intrapulmonary spread. J Thorac Oncol. 2009; 4:1490–5.
Article50. Eberhard DA, Giaccone G, Johnson BE; Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008; 26:983–94.
Article51. Nicholson AG, Gonzalez D, Shah P, et al. Refining the diagnosis and EGFR status of non-small cell lung carcinoma in biopsy and cytologic material, using a panel of mucin staining, TTF-1, cytokeratin 5/6, and P63, and EGFR mutation analysis. J Thorac Oncol. 2010; 5:436–41.
Article52. Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016; 2:1014–22.53. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016; 34:3375–82.
Article54. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013; 13:772–87.
Article55. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014; 5:4846.
Article56. Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014; 371:2167–77.
Article57. Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014; 371:1963–71.58. Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012; 18:375–7.59. Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012; 30:4352–9.60. Drilon A, Wang L, Hasanovic A, et al. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013; 3:630–5.61. Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med. 2017; 5:42–50.62. Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013; 19:1469–72.63. Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014; 20:4107–14.
Article64. Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res. 2014; 20:3087–93.
Article65. Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013; 3:636–47.66. Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011; 6:466–72.67. Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One. 2014; 9:e103333.68. Lee SE, Lee B, Hong M, et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol. 2015; 28:468–79.69. Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010; 16:1561–71.70. Conklin CM, Craddock KJ, Have C, Laskin J, Couture C, Ionescu DN. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol. 2013; 8:45–51.71. Soda M, Isobe K, Inoue A, et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res. 2012; 18:5682–9.72. Betz BL, Dixon CA, Weigelin HC, Knoepp SM, Roh MH. The use of stained cytologic direct smears for ALK gene rearrangement analysis of lung adenocarcinoma. Cancer Cytopathol. 2013; 121:489–99.73. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010; 2:62ra93.
Article74. Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013; 31:731–7.
Article75. Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011; 6:e20351.76. Lim SH, Sun JM, Choi YL, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: a single-arm, phase 2 study. Cancer. 2016; 122:3024–31.77. Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014; 20:3299–309.
Article78. Pearson A, Smyth E, Babina IS, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016; 6:838–51.79. Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005; 97:643–55.
Article80. Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009; 4:e7464.
Article81. Lee Y, Shim HS, Park MS, et al. High EGFR gene copy number and skin rash as predictive markers for EGFR tyrosine kinase inhibitors in patients with advanced squamous cell lung carcinoma. Clin Cancer Res. 2012; 18:1760–8.82. Schildhaus HU, Schultheis AM, Rüschoff J, et al. MET amplification status in therapy-naive adeno- and squamous cell carcinomas of the lung. Clin Cancer Res. 2015; 21:907–15.83. Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011; 6:942–6.84. Zhang Y, Wang W, Wang Y, et al. Response to crizotinib observed in lung adenocarcinoma with MET copy number gain but without a high-level MET/CEP7 ratio, MET overexpression, orexon 14 splicing mutations. J Thorac Oncol. 2016; 11:e59–62.85. Mar N, Vredenburgh JJ, Wasser JS. Targeting HER2 in the treatment of non-small cell lung cancer. Lung Cancer. 2015; 87:220–5.
Article86. Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012; 2:922–33.87. Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012; 25:1473–80.88. Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013; 31:3997–4013.89. Dziadziuszko R, Wynes MW, Singh S, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol. 2012; 7:340–7.90. Gadgeel SM, Chen W, Cote ML, et al. Fibroblast growth factor receptor 1 amplification in non-small cell lung cancer by quantitative real-time PCR. PLoS One. 2013; 8:e79820.
Article91. Goke F, Franzen A, Menon R, et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest. 2012; 142:1020–6.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Utilization of Cytologic Fine-Needle Aspirates of Lung Cancer for Molecular Diagnostic Testing
- Molecular Pathology of Lung Cancer: Current Status and Future Directions
- Biomarker testing of cytology specimens in personalized medicine for lung cancer patients
- What’s new in molecular genetic pathology 2021: solid tumors and NGS panel selection
- Current Issues in Lung Cancer Pathology