Transl Clin Pharmacol.
2017 Sep;25(3):147-152. 10.12793/tcp.2017.25.3.147.
Prediction and visualization of CYP2D6 genotype-based phenotype using clustering algorithms
- Affiliations
-
- 1Department of Clinical Pharmacology, Inje University College of Medicine, Busan Paik Hospital, Busan 47392, Republic of Korea. phshinjg@gmail.com
- 2Department of Pharmacology and PharmacoGenomics Research Center, Inje University College of Medicine, Busan 47392, Republic of Korea.
- 3Department of Clinical Pharmacology and Therapeutics, Seoul National University College of Medicine, Seoul 03080, Republic of Korea.
- KMID: 2390935
- DOI: http://doi.org/10.12793/tcp.2017.25.3.147
Abstract
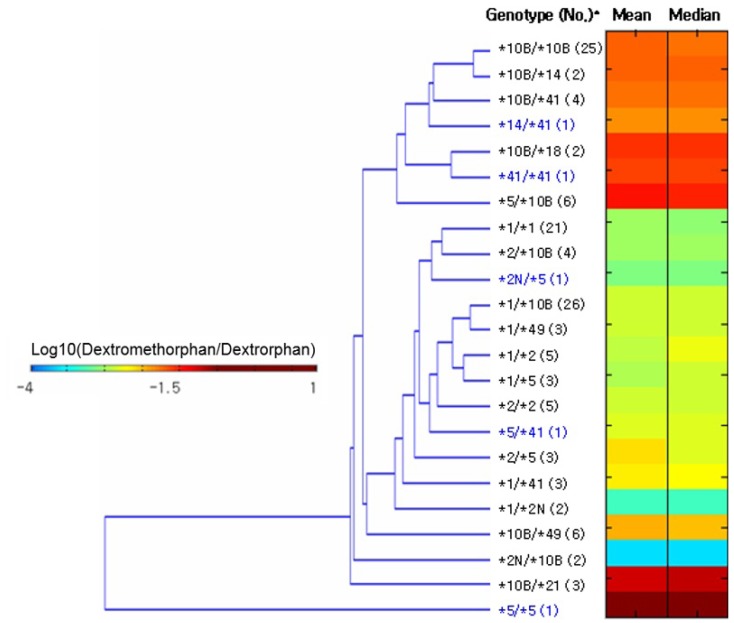
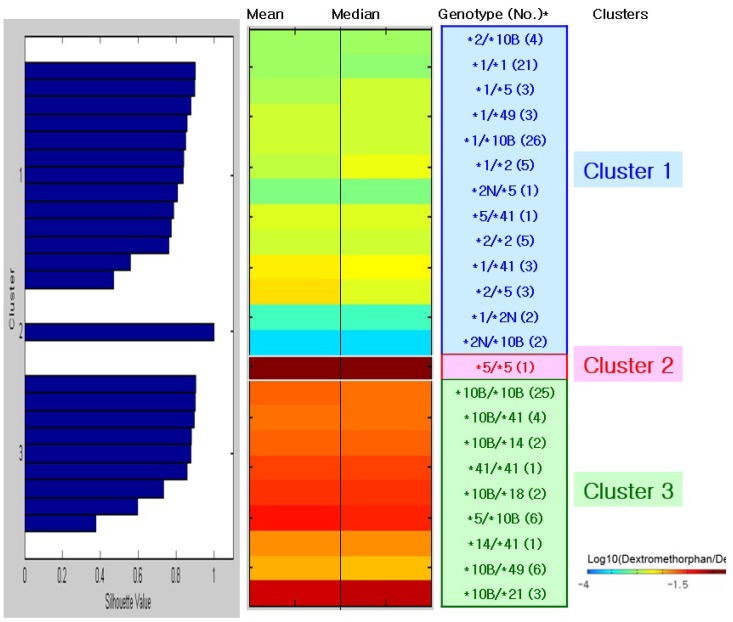
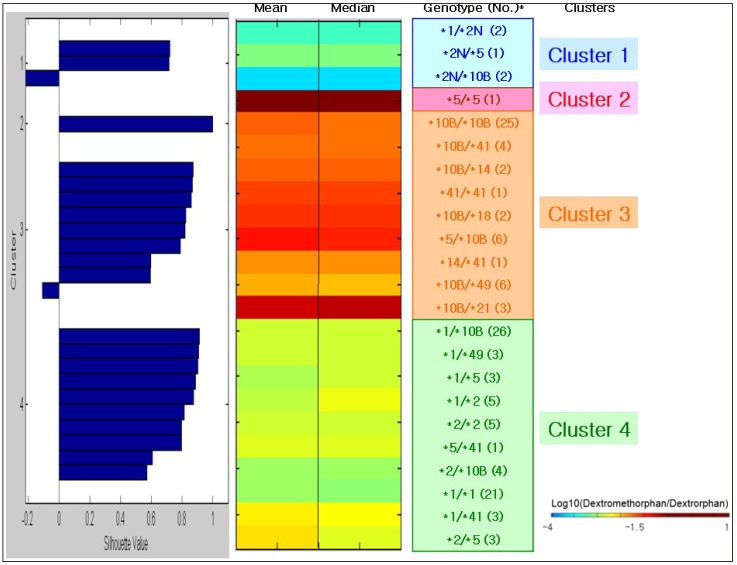
- This study focused on the role of cytochrome P450 2D6 (CYP2D6) genotypes to predict phenotypes in the metabolism of dextromethorphan. CYP2D6 genotypes and metabolic ratios (MRs) of dextromethorphan were determined in 201 Koreans. Unsupervised clustering algorithms, hierarchical and k-means clustering analysis, and color visualizations of CYP2D6 activity were performed on a subset of 130 subjects. A total of 23 different genotypes were identified, five of which were observed in one subject. Phenotype classifications were based on the means, medians, and standard deviations of the log MR values for each genotype. Color visualization was used to display the mean and median of each genotype as different color intensities. Cutoff values were determined using receiver operating characteristic curves from the k-means analysis, and the data were validated in the remaining subset of 71 subjects. Using the two highest silhouette values, the selected numbers of clusters were three (the best) and four. The findings from the two clustering algorithms were similar to those of other studies, classifying *5/*5 as a lowest activity group and genotypes containing duplicated alleles (i.e., CYP2D6*1/*2N) as a highest activity group. The validation of the k-means clustering results with data from the 71 subjects revealed relatively high concordance rates: 92.8% and 73.9% in three and four clusters, respectively. Additionally, color visualization allowed for rapid interpretation of results. Although the clustering approach to predict CYP2D6 phenotype from CYP2D6 genotype is not fully complete, it provides general information about the genotype to phenotype relationship, including rare genotypes with only one subject.
Keyword
MeSH Terms
Figure
Reference
-
1. Ingelman-Sundberg M, Oscarson M, McLellan RA. Polymorphic human cytochrome P450 enzymes: An opportunity for individualized drug treatment. Trends Pharmacol Sci. 1999; 20:342–349. PMID: 10431214.
Article2. Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med. 2007; 147:755–765. PMID: 18056659.
Article3. Ma MK, Woo MH, McLeod HL. Genetic basis of drug metabolism. Am J Health Syst Pharm. 2002; 59:2061–2069. PMID: 12434718.
Article4. Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013; 368:20120431. DOI: 10.1098/rstb.2012.0431. PMID: 23297354.
Article5. Cytochrome P450 homepage. html Accessed 18 July 2017. http://drnelson.uthsc.edu/cytochromeP450.6. Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005; 5:6–13. PMID: 15492763.
Article7. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013; 138:103–141. DOI: 10.1016/j.pharmthera.2012.12.007. PMID: 23333322.
Article8. Sim SC, Ingelman-Sundberg M. Update on allele nomenclature for human cytochromes P450 and the human cytochrome P450 allele (CYP-allele) nomenclature database. Methods Mol Biol. 2013; 987:251–259. DOI: 10.1007/978-1-62703-321-3_21. PMID: 23475683.
Article9. Bernard S, Neville KA, Nguyen AT, Flockhart DA. Interethnic differences in genetic polymorphisms of CYP2D6 in the U.S. Population: Clinical implications. Oncologist. 2006; 11:126–135. PMID: 16476833.
Article10. Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, et al. Standardizing terms for clinical pharmacogenetic test results: Consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017; 19:215–223. DOI: 10.1038/gim.2016.87. PMID: 27441996.
Article11. Lee SJ, Lee SS, Jung HJ, Kim HS, Park SJ, Yeo CW, et al. Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab Dispos. 2009; 37:1464–1470. DOI: 10.1124/dmd.108.022368. PMID: 19364831.12. Kim EY, Lee SS, Jung HJ, Jung HE, Yeo CW, Shon JH, et al. Robust CYP2D6 genotype assay including copy number variation using multiplex single-base extension for Asian populations. Clin Chim Acta. 2010; 411:2043–2048. DOI: 10.1016/j.cca.2010.08.042. PMID: 20828547.
Article13. Gaedigk A, Gotschall RR, Forbes NS, Simon SD, Kearns GL, Leeder JS. Optimization of cytochrome P450 2D6 (CYP2D6) phenotype assignment using a genotyping algorithm based on allele frequency data. Pharmacogenetics. 1999; 9:669–682. PMID: 10634130.14. Eichelbaum M, Woolhouse NM. Inter-ethnic difference in sparteine oxidation among Ghanaians and Germans. Eur J Clin Pharmacol. 1985; 28:79–83. PMID: 3987789.
Article15. Schmid B, Bircher J, Preisig R, Kupfer A. Polymorphic dextromethorphan metabolism: Co-segregation of oxidative O-demethylation with debrisoquin hydroxylation. Clin Pharmacol Ther. 1985; 38:618–624. PMID: 4064464.
Article16. Sachse C, Brockmöller J, Hildebrand M, Müller K, Roots I. Correctness of prediction of the CYP2D6 phenotype confirmed by genotyping 47 intermediate and poor metabolizers of debrisoquine. Pharmacogenetics. 1998; 8:181–185. PMID: 10022755.
Article17. Chou WH, Yan FX, Robbins-Weilert DK, Ryder TB, Liu WW, Perbost C, et al. Comparison of two CYP2D6 genotyping methods and assessment of genotype-phenotype relationships. Clin Chem. 2003; 49:542–551. PMID: 12651805.
Article18. Sabbagh A, Darlu P. Data-mining methods as useful tools for predicting individual drug response: Application to CYP2D6 data. Hum Hered. 2006; 62:119–134. PMID: 17057402.19. Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: Translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008; 83:234–242. PMID: 17971818.
Article20. Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: A critical assessment and call for standardization. Curr Drug Metab. 2014; 15:218–232. PMID: 24524666.
Article21. Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017; 19:69–76. DOI: 10.1038/gim.2016.80. PMID: 27388693.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- QCanvas: An Advanced Tool for Data Clustering and Visualization of Genomics Data
- Genetic polymorphisms of CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 in Vietnamese-Koreans
- CYP2D6 and NAT2 Polymorphism in HBV Associated in Hepatocellular Carcinoma Patients in Korea
- Selection of an Antidepressant Based on the Genotypes of Cytochrome P450 Enzymes Genes in a Patient with Major Depressive Disorder
- A Case Report of a Poor Metabolizer of CYP2D6 Presented with Unusual Responses to Nortriptyline Medication