J Korean Med Sci.
2017 Nov;32(11):1784-1791. 10.3346/jkms.2017.32.11.1784.
GAB2 Amplification in Squamous Cell Lung Cancer of Non-Smokers
- Affiliations
-
- 1Clinical Research Center, Asan Institute of Life Science, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 2Department of Biomedical Informatics, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 3Department of Pulmonology and Critical Medicine, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea. ccm@amc.seoul.kr
- 4Department of Laboratory Medicine, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 5Department of Oncology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 6Department of Thoracic Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 7Department of Pathology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 8Office of Clinical Research Information, Asan Institute of Life Science, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- KMID: 2390304
- DOI: http://doi.org/10.3346/jkms.2017.32.11.1784
Abstract
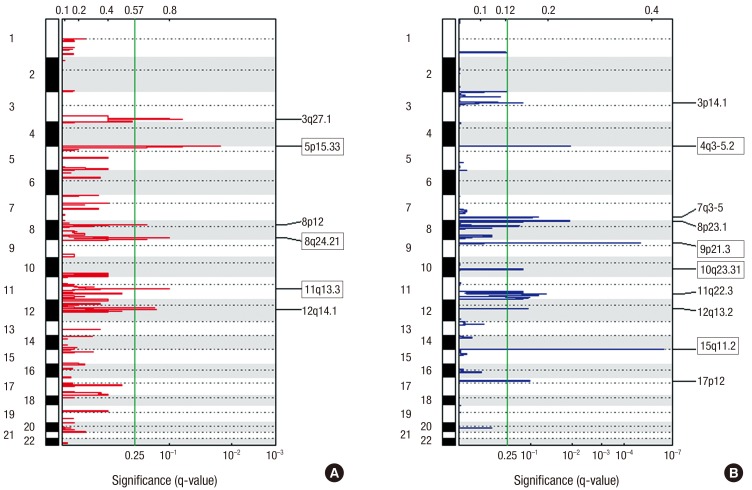
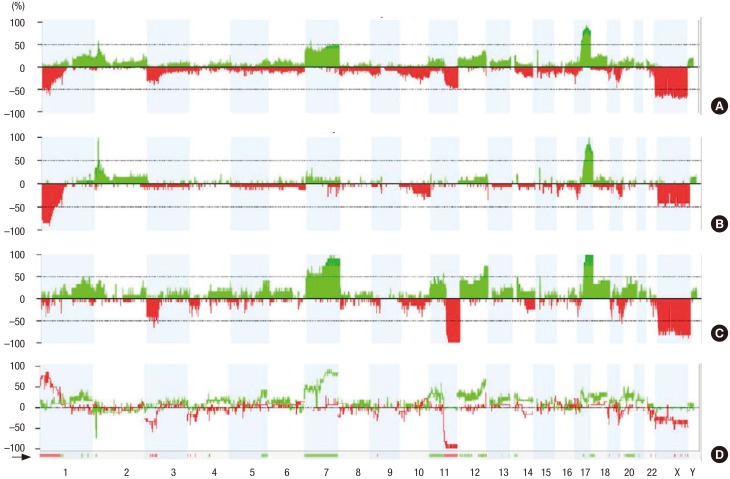
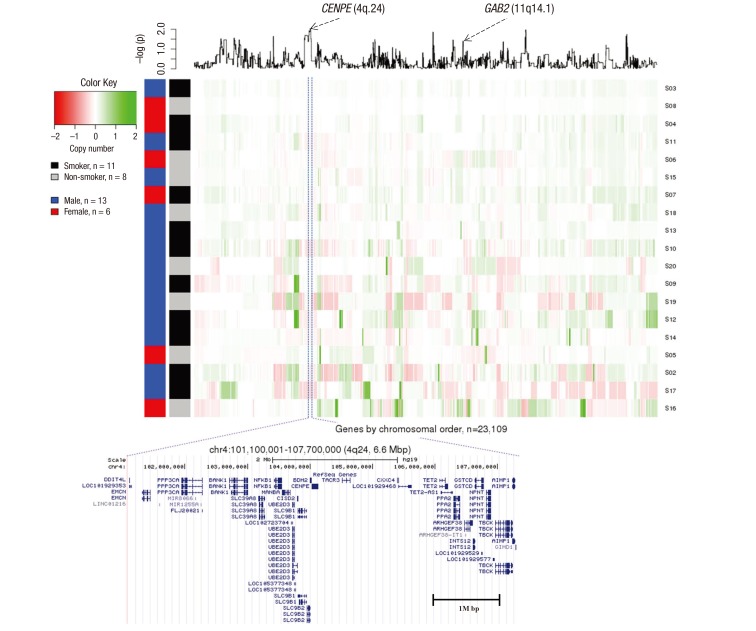
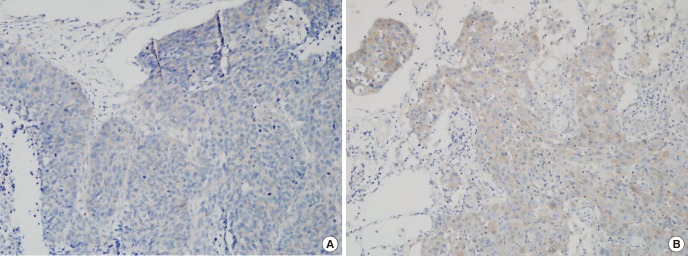
- Lung squamous cell cancer (SCC) is typically found in smokers and has a very low incidence in non-smokers, indicating differences in the tumor biology of lung SCC in smokers and non-smokers. However, the specific mutations that drive tumor growth in non-smokers have not been identified. To identify mutations in lung SCC of non-smokers, we performed a genetic analysis using arrays comparative genomic hybridization (ArrayCGH). We analyzed 19 patients with lung SCC who underwent surgical treatment between April 2005 and April 2015. Clinical characteristics were reviewed, and DNA was extracted from fresh frozen lung cancer specimens. All of copy number alterations from ArrayCGH were validated using The Cancer Genome Atlas (TCGA) copy number variation (CNV) data of lung SCC. We examined the frequency of copy number changes according to the smoking status (non-smoker [n = 8] or smoker [n = 11]). We identified 16 significantly altered regions from ArrayCGH data, three gain and four loss regions overlapped with the TCGA lung squamous cell carcinoma (LUSC) patients. Within these overlapped significant regions, we detected 15 genes that have been reported in the Cancer Gene census. We also found that the proto-oncogene GAB2 (11q14.1) was significantly amplified in non-smokers patients and vice versa in both ArrayCGH and TCGA data. Immunohistochemical analyses showed that GAB2 protein was relatively upregulated in non-smoker than smoker tissues (37.5% vs. 9.0%, P = 0.007). GAB2 amplification may have an important role in the development of lung SCC in non-smokers. GAB2 may represent a potential biomarker for lung SCC in non-smokers.
Keyword
MeSH Terms
Figure
Reference
-
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013; 63:11–30. PMID: 23335087.2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–2917. PMID: 21351269.3. Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007; 25:561–570. PMID: 17290066.4. Thun MJ, Hannan LM, Adams-Campbell LL, Boffetta P, Buring JE, Feskanich D, Flanders WD, Jee SH, Katanoda K, Kolonel LN, et al. Lung cancer occurrence in never-smokers: an analysis of 13 cohorts and 22 cancer registry studies. PLoS Med. 2008; 5:e185. PMID: 18788891.5. Koo LC, Ho JH. Worldwide epidemiological patterns of lung cancer in nonsmokers. Int J Epidemiol. 1990; 19(Suppl 1):S14–S23. PMID: 2258272.6. Gao YT, Blot WJ, Zheng W, Ershow AG, Hsu CW, Levin LI, Zhang R, Fraumeni JF Jr. Lung cancer among Chinese women. Int J Cancer. 1987; 40:604–609. PMID: 2824385.7. Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004; 101:13306–13311. PMID: 15329413.8. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004; 350:2129–2139. PMID: 15118073.9. Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004; 304:1497–1500. PMID: 15118125.10. Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest. 2004; 126:347–351. PMID: 15302716.11. Brunelli M, Bria E, Nottegar A, Cingarlini S, Simionato F, Caliò A, Eccher A, Parolini C, Iannucci A, Gilioli E, et al. True 3q chromosomal amplification in squamous cell lung carcinoma by FISH and aCGH molecular analysis: impact on targeted drugs. PLoS One. 2012; 7:e49689. PMID: 23236352.12. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. PMID: 22960745.13. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. PMID: 25605792.14. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011; 12:R41. PMID: 21527027.15. Ding C, Luo J, Fan X, Li L, Li S, Wen K, Feng J, Wu G. Elevated GAB2 induces tumor growth and angiogenesis in colorectal cancer through upregulating VEGF levels. J Exp Clin Cancer Res. 2017; 36:56–67. PMID: 28420432.16. Broad Institute TCGA Genome Data Analysis Center (US). SNP6 copy number analysis (GISTIC2) [Internet]. accessed on 9 September 2016. Available at http://gdac.broadinstitute.org/runs/analyses__2015_04_02/reports/cancer/LUSC/CopyNumber_Gistic2/nozzle.html.17. Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. PMID: 25355519.18. Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009; 4:e4653. PMID: 19247474.19. Minna JD. Molecular biology overview. In : Pass HI, Mitchell JB, Johnson DH, Turrisi AT, editors. Lung Cancer: Principles and Practice. Philadelphia, PA: Lippincott-Raven;1996. p. 143–148.20. Wöhrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009; 7:22. PMID: 19737390.21. Bentires-Alj M, Gil SG, Chan R, Wang ZC, Wang Y, Imanaka N, Harris LN, Richardson A, Neel BG, Gu H. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006; 12:114–121. PMID: 16369543.22. Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H. GAB2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene. 2012; 31:2512–2520. PMID: 21996746.23. Shi L, Sun X, Zhang J, Zhao C, Li H, Liu Z, Fang C, Wang X, Zhao C, Zhang X, et al. GAB2 expression in glioma and its implications for tumor invasion. Acta Oncol. 2013; 52:1739–1750. PMID: 23231021.24. Lee SH, Jeong EG, Nam SW, Lee JY, Yoo NJ, Lee SH. Increased expression of GAB2, a scaffolding adaptor of the tyrosine kinase signalling, in gastric carcinomas. Pathology. 2007; 39:326–329. PMID: 17558859.25. Chernoff KA, Bordone L, Horst B, Simon K, Twadell W, Lee K, Cohen JA, Wang S, Silvers DN, Brunner G, et al. GAB2 amplifications refine molecular classification of melanoma. Clin Cancer Res. 2009; 15:4288–4291. PMID: 19509136.26. Xu XL, Wang X, Chen ZL, Jin M, Yang W, Zhao GF, Li JW. Overexpression of Grb2-associated binder 2 in human lung cancer. Int J Biol Sci. 2011; 7:496–504. PMID: 21552417.27. Dunn GP, Cheung HW, Agarwalla PK, Thomas S, Zektser Y, Karst AM, Boehm JS, Weir BA, Berlin AM, Zou L, et al. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proc Natl Acad Sci USA. 2014; 111:1102–1107. PMID: 24385586.28. Zhang X, Zhang Y, Tao B, Wang D, Cheng H, Wang K, Zhou R, Xie Q, Ke Y. Docking protein GAB2 regulates mucin expression and goblet cell hyperplasia through TYK2/STAT6 pathway. FASEB J. 2012; 26:4603–4613. PMID: 22859374.29. Ding CB, Yu WN, Feng JH, Luo JM. Structure and function of GAB2 and its role in cancer. [Review]. Mol Med Rep. 2015; 12:4007–4014. PMID: 26095858.30. Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009; 8:627–644. PMID: 19644473.31. Maulik G, Madhiwala P, Brooks S, Ma PC, Kijima T, Tibaldi EV, Schaefer E, Parmar K, Salgia R. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. J Cell Mol Med. 2002; 6:539–553. PMID: 12611639.32. Xu LJ, Wang YC, Lan HW, Li J, Xia T. Grb2-associated binder-2 gene promotes migration of non-small cell lung cancer cells via Akt signaling pathway. Am J Transl Res. 2016; 8:1208–1217. PMID: 27158407.33. Daly RJ, Gu H, Parmar J, Malaney S, Lyons RJ, Kairouz R, Head DR, Henshall SM, Neel BG, Sutherland RL. The docking protein GAB2 is overexpressed and estrogen regulated in human breast cancer. Oncogene. 2002; 21:5175–5181. PMID: 12140767.34. Shin A, Oh CM, Kim BW, Woo H, Won YJ, Lee JS. Lung cancer epidemiology in Korea. Cancer Res Treat. 2017; 49:616–626. PMID: 27669705.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Features of Advanced Non-Small Cell Lung Cancer in Never-Smokers
- Druggable Targets of Squamous Cell Lung Cancer
- The National Survey of Lung Cancer in Korea
- The change of squamous cell cancer antigen(SCC Ag) level as a tumor marker in squamous cell lung cancer
- Clinicopathologic Analysis of Epidermal Growth Factor Receptor Status in Non-small Cell Lung Cancer: Protein Expression, Gene Amplification and Survival Analysis