J Breast Cancer.
2017 Jun;20(2):132-141. 10.4048/jbc.2017.20.2.132.
Overexpression of Uridine-Cytidine Kinase 2 Correlates with Breast Cancer Progression and Poor Prognosis
- Affiliations
-
- 1Prenatal Diagnostic Center, Huzhou Women and Children Hospital, Huzhou, China.
- 2Department of Epidemiology and Biostatistics, School of Basic Medical Sciences, Zhejiang Chinese Medical University, Hangzhou, China. myy@zcmu.edu.cn
- 3Hangzhou Hope Biotechnology Company, Hangzhou, China.
- 4Biomarkers Development, California Cancer Institute, Sino-American Cancer Foundation, Temple City, USA.
- 5General Medicine Division, College of Medicine, Taipei Medical University, Taipei, Taiwan.
- 6Key Lab of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China.
- 7Integrated Laboratory, Center of Translational Medicine, Taipei Medical University, Taipei, Taiwan.
- KMID: 2389750
- DOI: http://doi.org/10.4048/jbc.2017.20.2.132
Abstract
- PURPOSE
Uridine-cytidine kinase (UCK) 2 is a rate-limiting enzyme involved in the salvage pathway of pyrimidine-nucleotide biosynthesis. Recent studies have shown that UCK2 is overexpressed in many types of cancer and may play a crucial role in activating antitumor prodrugs in human cancer cells. In the current study, we evaluated the potential prognostic value of UCK2 in breast cancer.
METHODS
We searched public databases to explore associations between UCK2 gene expression and clinical parameters in patients with breast cancer. Gene set enrichment analysis (GSEA) was performed to identify biological pathways associated with UCK2 gene expression levels. Survival analyses were performed using 10 independent large-scale breast cancer microarray datasets.
RESULTS
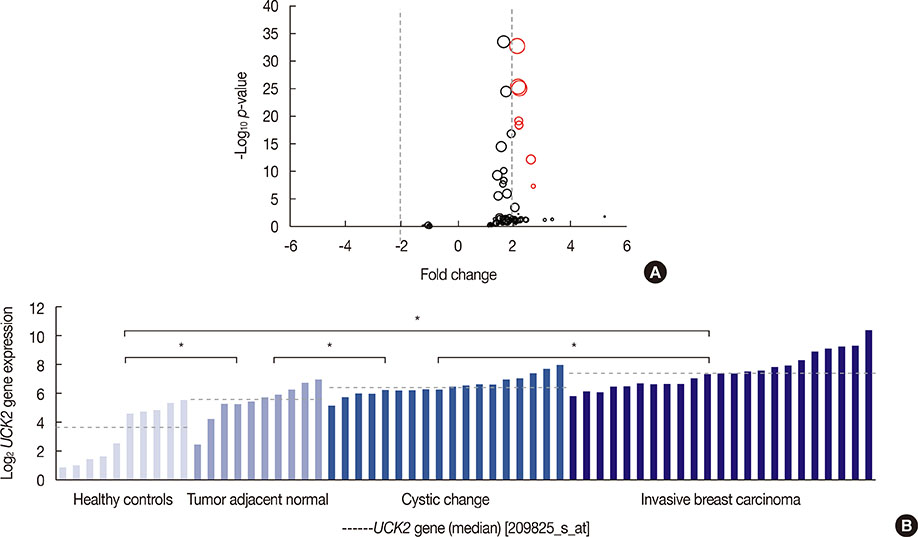
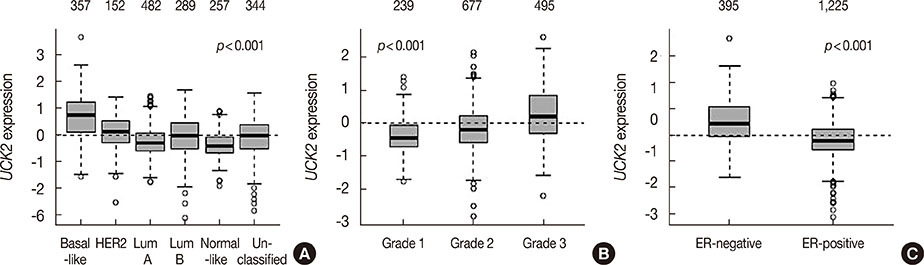
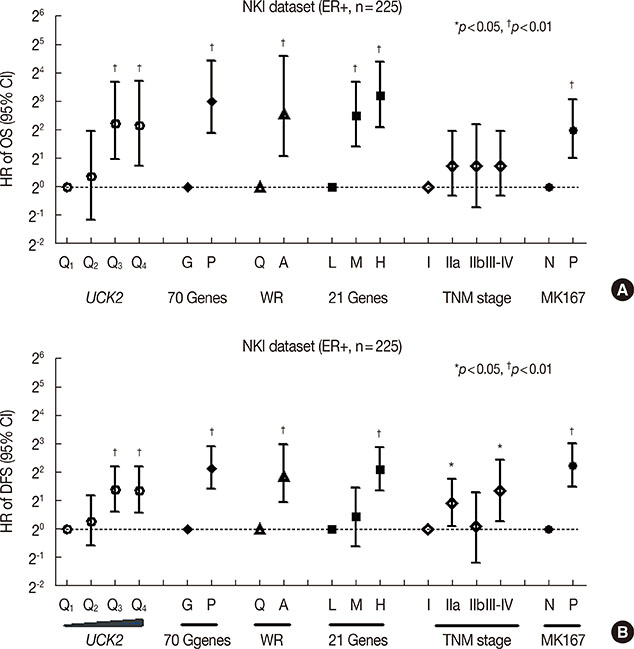
We found that UCK2 mRNA expression was elevated in breast cancer tissue compared with adjacent nontumorous tissue or breast tissue from healthy controls. High UCK2 levels were correlated with estrogen receptor negativity (p<0.001), advanced tumor grade (p<0.001), and poor tumor differentiation (p<0.001). GSEA revealed that UCK2-high breast cancers were enriched for gene sets associated with metastasis, progenitor-like phenotypes, and poor prognosis. Multivariable Cox proportional hazards regression analyses of microarray datasets verified that high UCK2 gene expression was associated with poor overall survival in a dose-response manner. The prognostic power of UCK2 was superior to that of TNM staging and comparable to that of multiple gene signatures.
CONCLUSION
These findings suggest that UCK2 may be a promising prognostic biomarker for patients with breast cancer.
MeSH Terms
Figure
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.
Article2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65:5–29.
Article3. André F, Domont J, Delaloge S. What can breast cancer molecular subclassification add to conventional diagnostic tools? Ann Oncol. 2007; 18:Suppl 9. ix33–ix36.
Article4. Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005; 23:7350–7360.
Article5. Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005; 353:1673–1684.
Article6. van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415:530–536.
Article7. Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sørlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005; 102:3738–3743.
Article8. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004; 351:2817–2826.
Article9. Van Rompay AR, Norda A, Lindén K, Johansson M, Karlsson A. Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol Pharmacol. 2001; 59:1181–1186.
Article10. Shimamoto Y, Koizumi K, Okabe H, Kazuno H, Murakami Y, Nakagawa F, et al. Sensitivity of human cancer cells to the new anticancer ribo-nucleoside TAS-106 is correlated with expression of uridine-cytidine kinase 2. Jpn J Cancer Res. 2002; 93:825–833.
Article11. Shen F, Look KY, Yeh YA, Weber G. Increased uridine kinase (ATP: uridine 5’-phosphotransferase; EC 2.7.1.48) activity in human and rat tumors. Cancer Biochem Biophys. 1998; 16:1–15.12. Davidsson J, Andersson A, Paulsson K, Heidenblad M, Isaksson M, Borg A, et al. Tiling resolution array comparative genomic hybridization, expression and methylation analyses of dup(1q) in Burkitt lymphomas and pediatric high hyperdiploid acute lymphoblastic leukemias reveal clustered near-centromeric breakpoints and overexpression of genes in 1q22-32.3. Hum Mol Genet. 2007; 16:2215–2225.
Article13. Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008; 68:1786–1796.
Article14. Ringnér M, Fredlund E, Häkkinen J, Borg Å, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011; 6:e17911.
Article15. Smeds J, Miller LD, Bjöhle J, Hall P, Klaar S, Liu ET, et al. Gene profile and response to treatment. Ann Oncol. 2005; 16:Suppl 2. ii195–ii202.
Article16. Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Geneexpression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005; 365:671–679.
Article17. Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006; 66:10292–10301.
Article18. Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong time dependence of the 76-gene prognostic signature for nodenegative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007; 13:3207–3214.
Article19. van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002; 347:1999–2009.
Article20. Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009; 69:4116–4124.
Article21. Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res Treat. 2012; 132:1049–1062.
Article22. Heikkinen T, Greco D, Pelttari LM, Tommiska J, Vahteristo P, Heikkilä P, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res. 2011; 13:R130.
Article23. Itoh M, Iwamoto T, Matsuoka J, Nogami T, Motoki T, Shien T, et al. Estrogen receptor (ER) mRNA expression and molecular subtype distribution in ER-negative/progesterone receptor-positive breast cancers. Breast Cancer Res Treat. 2014; 143:403–409.
Article24. Azim HA Jr, Brohée S, Peccatori FA, Desmedt C, Loi S, Lambrechts D, et al. Biology of breast cancer during pregnancy using genomic profiling. Endocr Relat Cancer. 2014; 21:545–554.
Article25. Jézéquel P, Loussouarn D, Guérin-Charbonnel C, Campion L, Vanier A, Gouraud W, et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015; 17:43.
Article26. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550.
Article27. Murata D, Endo Y, Obata T, Sakamoto K, Syouji Y, Kadohira M, et al. A crucial role of uridine/cytidine kinase 2 in antitumor activity of 3’-ethynyl nucleosides. Drug Metab Dispos. 2004; 32:1178–1182.
Article28. Muhale FA, Wetmore BA, Thomas RS, McLeod HL. Systems pharmacology assessment of the 5-fluorouracil pathway. Pharmacogenomics. 2011; 12:341–350.
Article29. Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, Maurer M, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007; 104:7564–7569.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Cancer/Testis Antigen CT45A1 Promotes Transcription of Oncogenic Sulfatase-2 Gene in Breast Cancer Cells and Is Sensible Targets for Cancer Therapy
- Longer Survival in Patients with Breast Cancer with Cyclin D1 Over-Expression after Tumor Recurrence: Longer, but Occupied with Disease
- Downstream Neighbor of Son Overexpression is Associated With Breast Cancer Progression and a Poor Prognosis
- c-erbB-2 Oncoprotein Overexpression in Breast Cancer
- Overexpression of c-erbB2 and its Relationship with Chemotherapy in Breast Ccancer