Ann Lab Med.
2017 Sep;37(5):398-407. 10.3343/alm.2017.37.5.398.
Multidrug Resistance Mechanisms of Carbapenem Resistant Klebsiella pneumoniae Strains Isolated in Chongqing, China
- Affiliations
-
- 1Department of Clinical Laboratory, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China. 1309898173@qq.com
- KMID: 2383915
- DOI: http://doi.org/10.3343/alm.2017.37.5.398
Abstract
- BACKGROUND
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is considered a serious global threat. However, little is known regarding the multidrug resistance (MDR) mechanisms of CRKP. This study investigated the phenotypes and MDR mechanisms of CRKP and identified their clonal characteristics.
METHODS
PCR and sequencing were utilized to identify antibiotic resistance determinants. Integron gene cassette arrays were determined by restriction fragment length polymorphism (RFLP) analysis. Multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were used for epidemiological analysis. Plasmids were typed by using a PCR-based replicon typing and analyzed by conjugation and transformation assays.
RESULTS
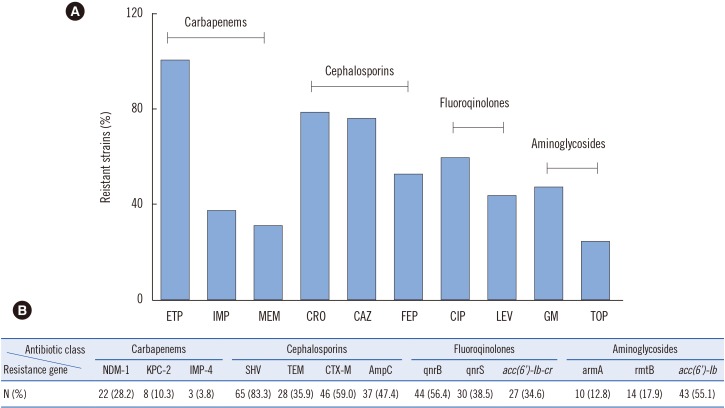
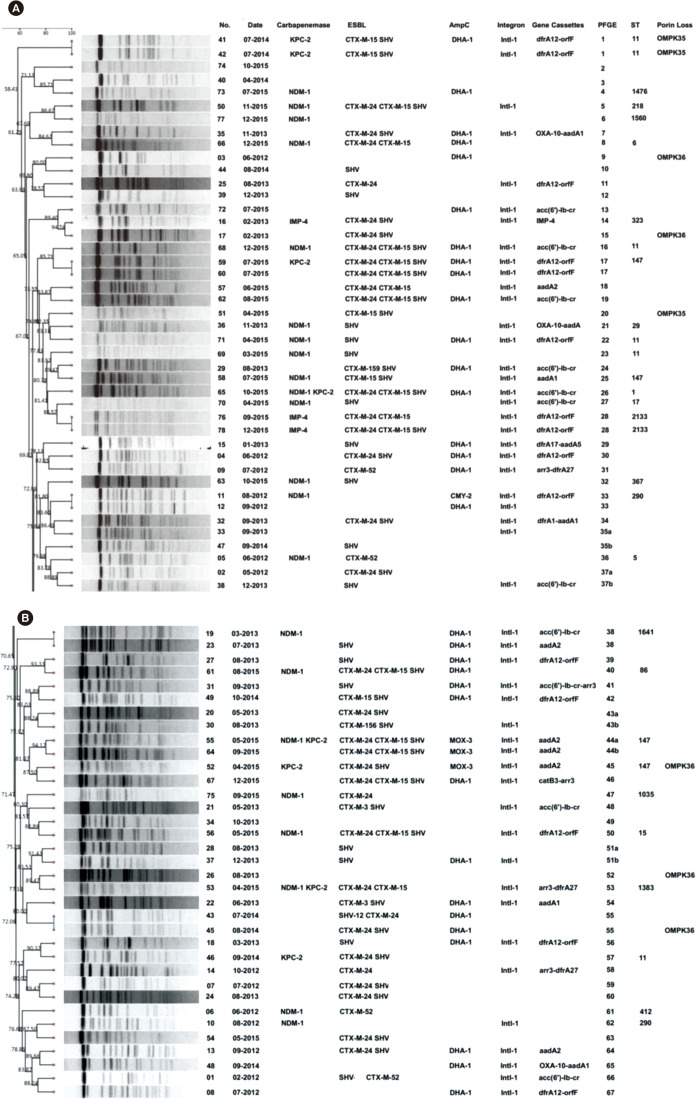
Seventy-eight strains were identified as resistant to at least one carbapenem; these CRKP strains had a high prevalence rate (38.5%, 30/78) of carbapenemase producers. Additionally, most isolates harbored MDR genes, including Extended spectrum β-lactamases (ESBLs), AmpC, and quinolone and aminoglycoside resistance genes. Loss of porin genes was observed, and Class 1 integron was detected in 66.7% of the investigated isolates. PFGE and MLST results excluded the occurrence of clonal dissemination among these isolates.
CONCLUSIONS
A high prevalence of NDM-1 genes encoding carbapenem resistance determinants was demonstrated among the K. pneumoniae isolates. Importantly, this is the first report of bla(NDM-1) carriage in a K. pneumoniae ST1383 clone in China and of a MDR CRKP isolate co-harboring bla(NDM-1), bla(KPC-2), bla(CTX-M), bla(SHV), acc(6"²)-Ib, rmtB, qnrB, and acc(6"²)-Ib-cr.
Keyword
MeSH Terms
-
China*
Clone Cells
Drug Resistance, Bacterial
Drug Resistance, Microbial
Drug Resistance, Multiple*
Electrophoresis, Gel, Pulsed-Field
Genes, MDR
Integrons
Klebsiella pneumoniae*
Klebsiella*
Molecular Epidemiology
Phenotype
Plasmids
Pneumonia
Polymerase Chain Reaction
Polymorphism, Restriction Fragment Length
Prevalence
Replicon
Figure
Reference
-
1. Hawser SP, Bouchillon SK, Lascols C, Hackel M, Hoban DJ, Badal RE, et al. Susceptibility of Klebsiella pneumoniae isolates from intra-abdominal infections and molecular characterization of ertapenem-resistant isolates. Antimicrob Agents Chemother. 2011; 55:3917–3921. PMID: 21670192.2. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009; 9:228–236. PMID: 19324295.3. Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class a carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009; 47:1631–1639. PMID: 19386850.4. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011; 66:307–312. PMID: 21131324.5. Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli Isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008; 52:2014–2018. PMID: 18332176.6. Zhou G, Guo S, Luo Y, Ye L, Song Y, Sun G, et al. NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg Infect Dis. 2014; 20:340–342. PMID: 24456600.7. Zheng R, Zhang Q, Guo Y, Feng Y, Liu L, Zhang A, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016; 15:10. PMID: 26896089.8. Jin Y, Shao C, Li J, Fan H, Bai Y, Wang Y. Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS One. 2015; 10:e0119571. PMID: 25799421.9. Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, et al. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother. 2014; 58:4275–4282. PMID: 24777095.10. Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Tests for Bacteria that Grow Aerobically. Approved Standard, 10th ed. M07-A10. Wayne, PA: Clinical and Laboratory Standards Institute;2015.11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 23th informational supplement, M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute;2014.12. Zhang C, Xu X, Pu S, Huang S, Sun J, Yang S, et al. Characterization of carbapenemases, extended spectrum β-lactamases, quinolone resistance and aminoglycoside resistance determinants in carbapenem-non-susceptible Escherichia coli from a teaching hospital in Chongqing, Southwest China. Infect Genet Evol. 2014; 27:271–276. PMID: 25107431.13. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009; 63:659–667. PMID: 19233898.14. Bado I, Cordeiro NF, Robino L, García-Fulgueiras V, Seija V, Bazet C, et al. Detection of class 1 and 2 integrons, extended-spectrum β-lactamases and qnr alleles in enterobacterial isolates from the digestive tract of intensive care unit inpatients. Int J Antimicrob Agents. 2010; 36:453–458. PMID: 20691572.15. Correia M, Boavida F, Grosso F, Salgado MJ, Lito LM, Cristino JM, et al. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2003; 47:2838–2843. PMID: 12936982.16. Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995; 39:185–191. PMID: 7695304.17. Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, et al. Outbreak of carbapenem-resistant Enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis. 2012; 55:e109–e117. PMID: 22997214.18. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005; 63:219–228. PMID: 15935499.19. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–2239. PMID: 7494007.20. Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2011; 55:1485–1493. PMID: 21282452.21. Kim SY, Shin J, Shin SY, Ko KS. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis. 2013; 76:486–490. PMID: 23688521.22. Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, et al. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 2009; 63:427–437. PMID: 19155227.23. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010; 10:597–602. PMID: 20705517.24. Ranjan A, Shaik S, Mondal A, Nandanwar N, Hussain A, Semmler T, et al. Molecular epidemiology and genome dynamics of New Delhi Metallo-β-Lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob Agents Chemother. 2016; 60:6795–6805. PMID: 27600040.25. Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, Al Jindan RY, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014; 58:3085–3090. PMID: 24637692.26. Gharout-Sait A, Alsharapy SA, Brasme L, Touati A, Kermas R, Bakour S, et al. Enterobacteriaceae isolates carrying the New Delhi mettallo-β-lactamase gene in Yemen. J Med Microbiol. 2014; 63:1316–1323. PMID: 25009193.27. Tijet N, Sheth PM, Lastovetska O, Chung C, Patel SN, Melano RG. Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008-2011. PLoS One. 2014; 9:e116421. PMID: 25549365.28. Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis. 2014; 28:186–192. PMID: 25245001.29. Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009; 64(S1):i3–i10. PMID: 19675017.30. Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014; 2:PLAS-0006-2013.31. Rodríguez Martínez JM, Díaz-de Alba P, Lopez-Cerero , Ruiz-Carrascoso G, Gomez-Gil R, Pascual A. Presence of quinolone resistance to qnrB1 genes and blaOXA-48 carbapenemase in clinical isolates of Klebsiella pneumoniae in Spain. Enferm Infecc Microbiol Clin. 2014; 32:441–442. PMID: 24746402.32. Ruiz E, Sáenz Y, Zarazaga M, Rocha-Gracia R, Martínez-Martínez L, Arlet G, Torres C. qnr, aac(6)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: genetic environments and plasmid and chromosomal location. J Antimicrob Chemother. 2012; 67:886–897. PMID: 22223228.33. Fendukly F, Karlsson I, Hanson HS, Kronvall G, Dornbusch K. Patterns of mutations in target genes in septicemia isolates of Escherichia coli and Klebsiella pneumoniae with resistance or reduced susceptibility to ciprofloxacin. APMIS. 2003; 111:857–866. PMID: 14510643.34. Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007; 51:3354–3360. PMID: 17548499.35. Almaghrabi R, Clancy CJ, Doi Y, Hao B, Chen L, Shields RK, et al. Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob Agents Chemother. 2014; 58:4443–4451. PMID: 24867988.36. Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2006; 50:3861–3866. PMID: 16940067.37. Leverstein-van Hall MA, M Blok HE, T Donders AR, Paauw A, Fluit AC, Verhoef J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis. 2003; 187:251–259. PMID: 12552449.38. Wu W, Feng Y, Carattoli A, Zong Z. Characterization of an Enterobacter cloacae strain producing both KPC and NDM carbapenemases by whole-genome sequencing. Antimicrob Agents Chemother. 2015; 59:6625–6628. PMID: 26248381.39. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009; 22:664–689. PMID: 19822894.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antimicrobial Therapy for Infections Caused by Carbapenem-Resistant Gram-Negative Bacteria
- Plasmid-Mediated Resistance to Extended-Spectrum beta-lactams in Enterobacter cloacae: Report of 4 cases
- Characterization of Carbapenemase Genes in Enterobacteriaceae Species Exhibiting Decreased Susceptibility to Carbapenems in a University Hospital in Chongqing, China
- Phenotypic and Genomic Characterization of AmpC-Producing Klebsiella pneumoniae From Korea
- Analysis of blood culture data in Korea: bacterial distribution and cumulative antimicrobial resistance (2016–2020)