Proposal of an Appropriate Decalcification Method of Bone Marrow Biopsy Specimens in the Era of Expanding Genetic Molecular Study
- Affiliations
-
- 1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea. revita@naver.com
- KMID: 2381382
- DOI: http://doi.org/10.4132/jptm.2015.03.16
Abstract
- BACKGROUND
The conventional method for decalcification of bone specimens uses hydrochloric acid (HCl) and is notorious for damaging cellular RNA, DNA, and proteins, thus complicating molecular and immunohistochemical analyses. A method that can effectively decalcify while preserving genetic material is necessary.
METHODS
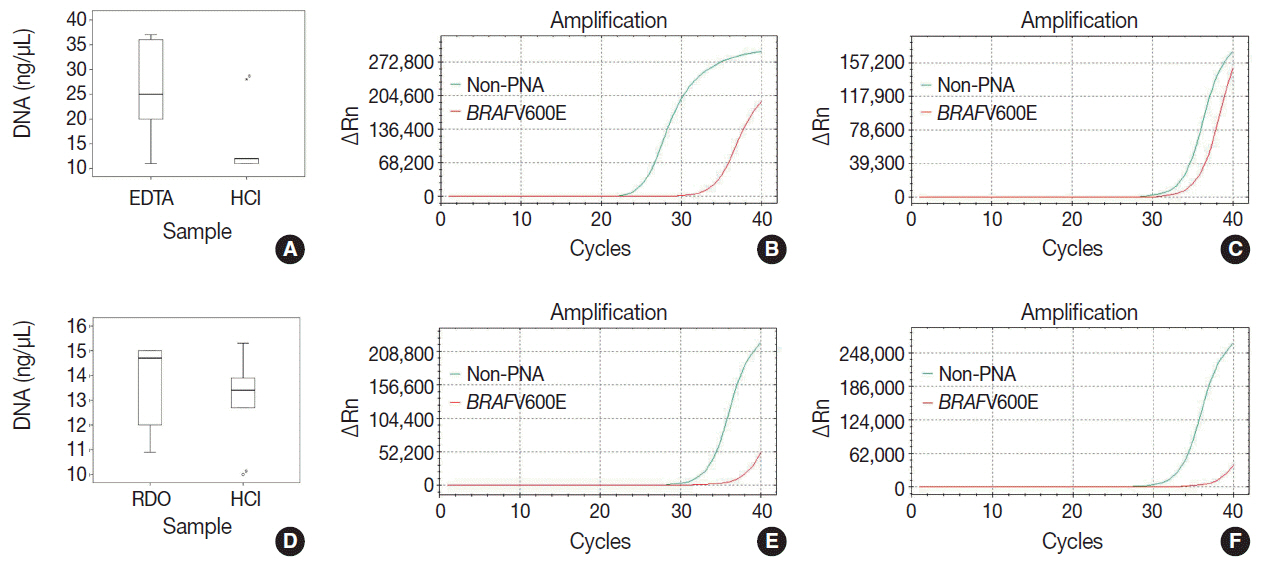
Pairs of bilateral bone marrow biopsies sampled from 53 patients were decalcified according to protocols of two comparison groups: EDTA versus HCl and RDO GOLD (RDO) versus HCl. Pairs of right and left bone marrow biopsy samples harvested from 28 cases were allocated into the EDTA versus HCl comparison group, and 25 cases to the RDO versus HCl comparison group. The decalcification protocols were compared with regards to histomorphology, immunohistochemistry, and molecular analysis. For molecular analysis, we randomly selected 5 cases from the EDTA versus HCl and RDO versus HCl groups.
RESULTS
The decalcification time for appropriate histomorphologic analysis was the longest in the EDTA method and the shortest in the RDO method. EDTA was superior to RDO or HCl in DNA yield and integrity, assessed via DNA extraction, polymerase chain reaction, and silver in situ hybridization using DNA probes. The EDTA method maintained intact nuclear protein staining on immunohistochemistry, while the HCl method produced poor quality images. Staining after the RDO method had equivocal results. RNA in situ hybridization using kappa and lambda RNA probes measured RNA integrity; the EDTA and RDO method had the best quality, followed by HCl.
CONCLUSIONS
The EDTA protocol would be the best in preserving genetic material. RDO may be an acceptable alternative when rapid decalcification is necessary.
Keyword
MeSH Terms
Figure
Cited by 3 articles
-
Extremely Well-Differentiated Papillary Thyroid Carcinoma Resembling Adenomatous Hyperplasia Can Metastasize to the Skull: A Case Report
Ju Yeon Pyo, Jisup Kim, Sung-eun Choi, Eunah Shin, Seok-Woo Yang, Cheong Soo Park, Seok-Mo Kim, SoonWon Hong
Yonsei Med J. 2017;58(1):255-258. doi: 10.3349/ymj.2017.58.1.255.Good Laboratory Standards for Clinical Next-Generation Sequencing Cancer Panel Tests
Jihun Kim, Woong-Yang Park, Nayoung K. D. Kim, Se Jin Jang, Sung-Min Chun, Chang-Ohk Sung, Jene Choi, Young-Hyeh Ko, Yoon-La Choi, Hyo Sup Shim, Jae-Kyung Won
J Pathol Transl Med. 2017;51(3):191-204. doi: 10.4132/jptm.2017.03.14.Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
J Pathol Transl Med. 2021;55(3):181-191. doi: 10.4132/jptm.2021.03.23.
Reference
-
1. Singh VM, Salunga RC, Huang VJ, et al. Analysis of the effect of various decalcification agents on the quantity and quality of nucleic acid (DNA and RNA) recovered from bone biopsies. Ann Diagn Pathol. 2013; 17:322–6.
Article2. Reineke T, Jenni B, Abdou MT, et al. Ultrasonic decalcification offers new perspectives for rapid FISH, DNA, and RT-PCR analysis in bone marrow trephines. Am J Surg Pathol. 2006; 30:892–6.
Article3. Brown RS, Edwards J, Bartlett JW, Jones C, Dogan A. Routine acid decalcification of bone marrow samples can preserve DNA for FISH and CGH studies in metastatic prostate cancer. J Histochem Cytochem. 2002; 50:113–5.
Article4. Alers JC, Krijtenburg PJ, Vissers KJ, van Dekken H. Effect of bone decalcification procedures on DNA in situ hybridization and comparative genomic hybridization. EDTA is highly preferable to a routinely used acid decalcifier. J Histochem Cytochem. 1999; 47:703–10.
Article5. Adegboyega PA, Gokhale S. Effect of decalcification on the immunohistochemical expression of ABH blood group isoantigens. Appl Immunohistochem Mol Morphol. 2003; 11:194–7.
Article6. Castania VA, Silveira JW, Issy AC, et al. Advantages of a combined method of decalcification compared to EDTA. Microsc Res Tech. 2015; 78:111–8.
Article7. Wickham CL, Sarsfield P, Joyner MV, Jones DB, Ellard S, Wilkins B. Formic acid decalcification of bone marrow trephines degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol. 2000; 53:336.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- An Experimental Study for Minimum Level of Decalcification to Detect the Osteolytic Bone Metastasis of Long Bone on Plain Radiography
- Bone Marrow Examination: Adventures in Diagnostic Hematology
- An Effective Role Pulsed Unipolar Magnetic Field for Bony Decalcification
- A case of bone marrow necrosis due to miliary tuberculosis
- Evaluation for usefulness of bone marrow study in fever of unknown origin