Ann Lab Med.
2017 Jul;37(4):331-335. 10.3343/alm.2017.37.4.331.
Detection of Immunoglobulin Heavy Chain Gene Clonality by Next-Generation Sequencing for Minimal Residual Disease Monitoring in B-Lymphoblastic Leukemia
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea. LEE.ST@yuhs.ac CJR0606@yuhs.ac
- 2Department of Laboratory Medicine, Hallym University College of Medicine, Kangnam Sacred Heart Hospital, Seoul, Korea.
- 3Brain Korea 21 PLUS Project for Medical Science, Yonsei University, Seoul, Korea.
- KMID: 2376791
- DOI: http://doi.org/10.3343/alm.2017.37.4.331
Abstract
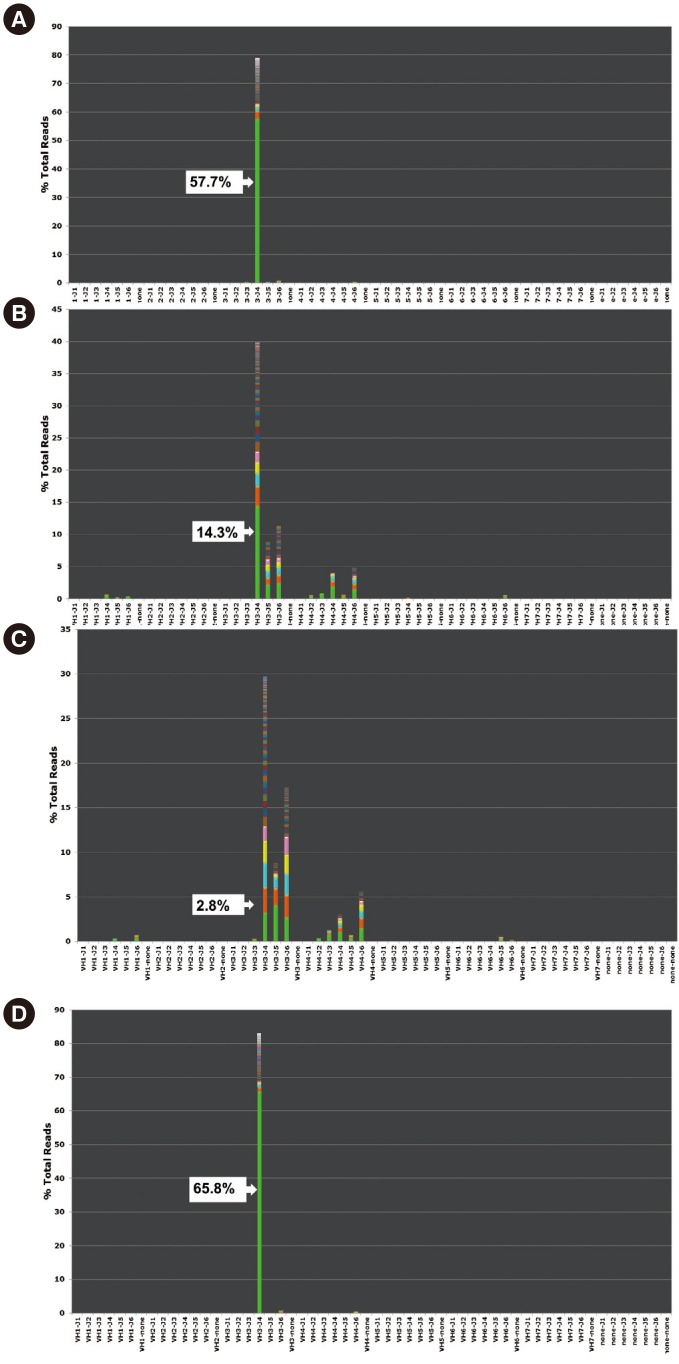
- Minimal residual disease (MRD) following B-lymphoblastic leukemia (B-ALL) treatment has gained prognostic importance. Clonal immunoglobulin heavy chain (IGH) gene rearrangement is a useful follow-up marker in B-ALL owing to its high positivity rate. We evaluated the performance and clinical applicability of a next-generation sequencing (NGS) assay for IGH rearrangement in B-ALL MRD monitoring. IGH rearrangement was tested by using fluorescence PCR-fragment analysis and the NGS assay in eight B-ALL patients. The NGS assay was run on two platforms: the Ion Torrent PGM (Thermo Fisher Scientific, USA) (18 samples from 1st to 7th patients) and the MiSeq system (Illumina, USA) (four samples from 8th patient). All initial diagnostic samples and four follow-up samples were positive for clonal IGH rearrangement with fluorescence PCR-fragment analysis and the NGS assay, and six follow-up samples were positive only with NGS. In one case with BCR-ABL1 translocation, BCR-ABL1 quantitative PCR was negative but the NGS IGH assay was positive just prior to full-blown relapse, suggesting the high sensitivity and clinical utility of the NGS assay. The NGS assay is proposed for MRD monitoring in B-ALL Additional studies are needed to confirm the clinical implications of cases showing positive results only in NGS.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
Minimal residual disease in acute lymphoblastic leukemia: technical aspects and implications for clinical interpretation
In-Suk Kim
Blood Res. 2020;55(Supplement):S19-S26. doi: 10.5045/br.2020.S004.Minimal residual disease in acute lymphoblastic leukemia: technical aspects and implications for clinical interpretation
In-Suk Kim
Blood Res. 2020;55(S1):S19-S26. doi: 10.5045/br.2020.S004.
Reference
-
1. Borowitz MJ, Devidas M, Hunger SP, Bowman WP, Carroll AJ, Carroll WL, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children's Oncology Group study. Blood. 2008; 111:5477–5485. PMID: 18388178.2. Logan AC, Vashi N, Faham M, Carlton V, Kong K, Buño I, et al. Immunoglobulin and T cell receptor gene high-throughput sequencing quantifies minimal residual disease in acute lymphoblastic leukemia and predicts post-transplantation relapse and survival. Biol Blood Marrow Transplant. 2014; 20:1307–1313. PMID: 24769317.3. Bassan R, Spinelli O, Oldani E, Intermesoli T, Tosi M, Peruta B, et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL). Blood. 2009; 113:4153–4162. PMID: 19141862.4. Schwartz RS. Shattuck lecture: Diversity of the immune repertoire and immunoregulation. N Engl J Med. 2003; 348:1017–1026. PMID: 12637612.5. Grove CS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech. 2014; 7:941–951. PMID: 25056697.6. Wu D, Emerson RO, Sherwood A, Loh ML, Angiolillo A, Howie B, et al. Detection of minimal residual disease in B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin Cancer Res. 2014; 20:4540–4548. PMID: 24970842.7. Vij R, Mazumder A, Klinger M, O'Dea D, Paasch J, Martin T, et al. Deep sequencing reveals myeloma cells in peripheral blood in majority of multiple myeloma patients. Clin Lymphoma Myeloma Leuk. 2014; 14:131–139.e1. PMID: 24629890.8. He J, Wu J, Jiao Y, Wagner-Johnston N, Ambinder RF, Diaz LA Jr, et al. IgH gene rearrangements as plasma biomarkers in Non-Hodgkin's lymphoma patients. Oncotarget. 2011; 2:178–185. PMID: 21399237.9. Langerak AW, Groenen PJ, Jm van Krieken JH, van Dongen JJ. Immunoglobulin/T-cell receptor clonality diagnostics. Expert Opin Med Diagn. 2007; 1:451–461. PMID: 23496353.10. Singh RR, Patel KP, Routbort MJ, Reddy NG, Barkoh BA, Handal B, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013; 15:607–622. PMID: 23810757.11. Pulsipher MA, Carlson C, Langholz B, Wall DA, Schultz KR, Bunin N, et al. IgH-V(D)J NGS-MRD measurement pre- and early post-allotransplant defines very low- and very high-risk ALL patients. Blood. 2015; 125:3501–3508. PMID: 25862561.12. Faham M, Zheng J, Moorhead M, Carlton VE, Stow P, Coustan-Smith E, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012; 120:5173–5180. PMID: 23074282.13. Gawad C, Pepin F, Carlton VE, Klinger M, Logan AC, Miklos DB, et al. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood. 2012; 120:4407–4417. PMID: 22932801.14. Langerak AW, Groenen PJ, Jm van Krieken JH, van Dongen JJ. Immunoglobulin/T-cell receptor clonality diagnostics. Expert Opin Med Diagn. 2007; 1:451–461. PMID: 23496353.15. Ladetto M, Brüggemann M, Monitillo L, Ferrero S, Pepin F, Drandi D, et al. Next-generation sequencing and real-time quantitative PCR for minimal residual disease detection in B-cell disorders. Leukemia. 2014; 28:1299–1307. PMID: 24342950.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Minimal residual disease in acute lymphoblastic leukemia: technical aspects and implications for clinical interpretation
- Non-Radioactive Detection of Clonality in Malignant Lymphoid Neoplasms using the Polymerase Chain Reaction
- Minimal Residual Disease Detection in Pediatric Acute Lymphoblastic Leukemia
- Comparison of Measurable Residual Disease in Pediatric B-Lymphoblastic Leukemia Using Multiparametric Flow Cytometry and Next-Generation Sequencing
- Measurements of treatment response in childhood acute leukemia