Ann Lab Med.
2017 Jul;37(4):313-319. 10.3343/alm.2017.37.4.313.
Detection of Serum Antibodies to Hepatitis E Virus Based on HEV Genotype 3 ORF2 Capsid Protein Expressed in Nicotiana benthamiana
- Affiliations
-
- 1Department of Plant Physiology and Molecular Biology, University of Plovdiv “Paisii Hilendarskiâ€, Plovdiv, Bulgaria. gerganaz@uni-plovdiv.bg
- 2Department of Animal Genetics, Faculty of Veterinary Medicine, Trakia University, Stara Zagora, Bulgaria.
- 3National Centre of Infectious and Parasitic Diseases, Sofia, Bulgaria.
- 4Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, UK.
- KMID: 2376787
- DOI: http://doi.org/10.3343/alm.2017.37.4.313
Abstract
- BACKGROUND
Hepatitis E virus (HEV) causes epidemics in developing countries and is primarily transmitted through the fecal-oral route. There have been recent reports on the zoonotic spread of the virus, and several animal species, primarily pigs, have been recognized as reservoirs of HEV. Because of its possible spread, there is an urgent need of a method for the cost-effective production of HEV proteins that can be used as diagnostic antigens for the serological detection of anti-HEV antibodies.
METHODS
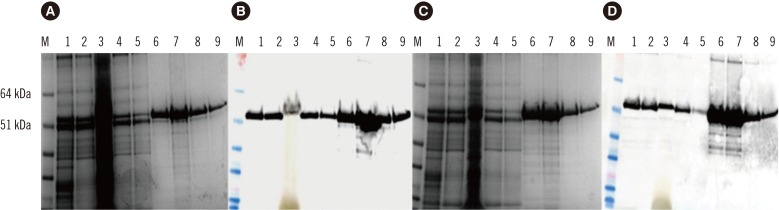
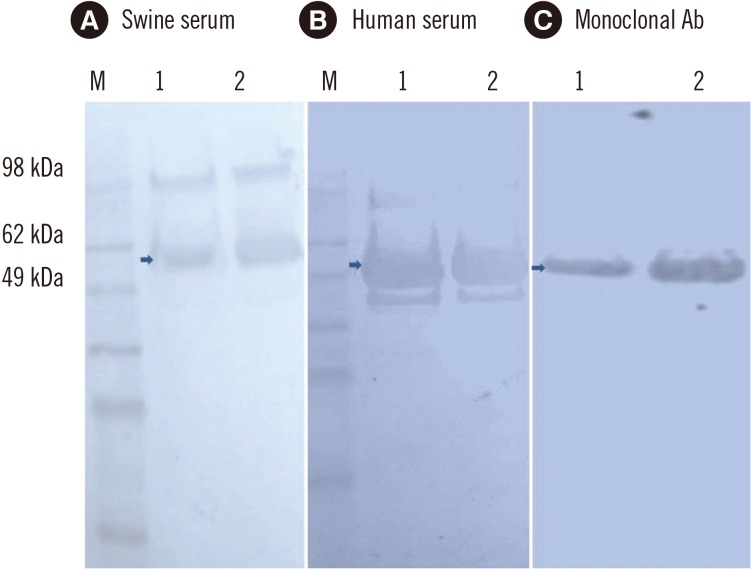
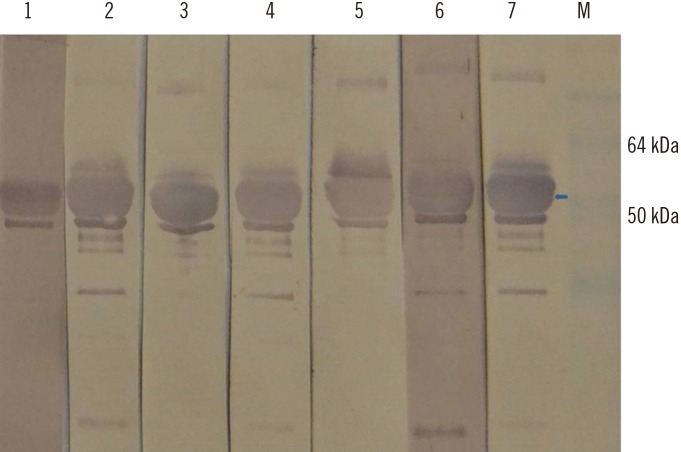
The HEV open reading frame (ORF)2 protein was purified from plant tissue by using immobilized metal-anion chromatography (IMAC). The recombinant protein was used to develop an in-house ELISA for testing anti-HEV antibodies in both human and swine sera. Thirty-six serum samples collected from patients with serologically proven HEV infection with commercial kits were tested for anti-HEV IgG antibodies by using the plant-expressed protein. Forty-five serum samples collected from apparently healthy pigs in Bulgarian farms were also tested.
RESULTS
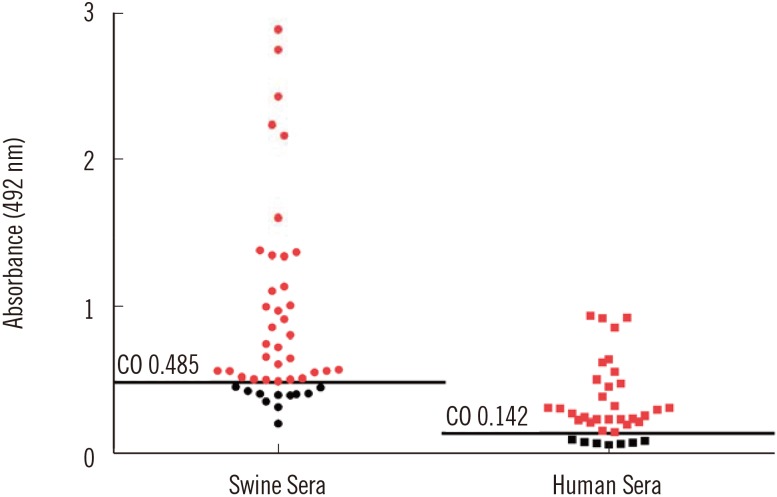
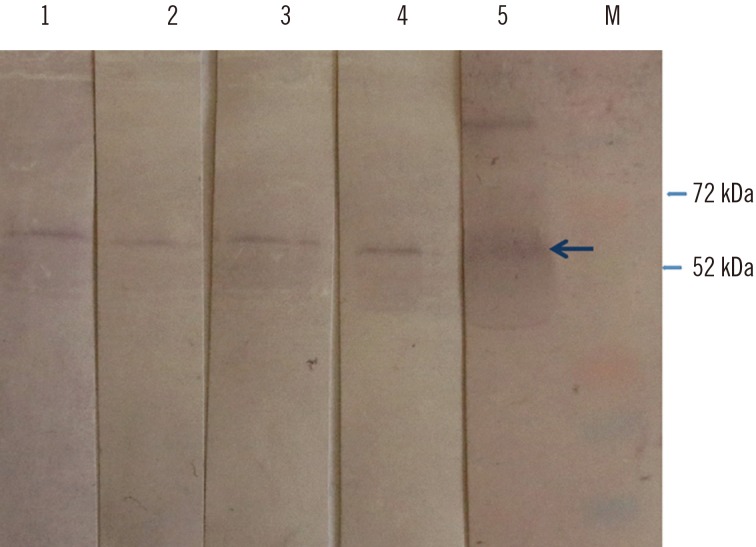
We confirmed the transient expression and purification of a truncated version of the HEV genotype 3 capsid protein in Nicotiana benthamiana and its usefulness as a diagnostic antigen. ELISA showed the presence of anti-HEV IgG antibodies in 29 of the 36 human samples. The in-house ELISA showed anti-HEV IgG antibodies in 34 of the 45 pigs.
CONCLUSIONS
We describe a method for the production of HEV ORF2 protein in N. benthamiana and the usefulness of this protein for the serological detection of anti-HEV antibodies in both humans and swine.
MeSH Terms
Figure
Reference
-
1. Meng XJ. Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis. 2013; 33:41–49. PMID: 23564388.2. Di Bartolo I, Ponterio E, Castellini L, Ostanello F, Ruggeri FM. Viral and antibody HEV prevalence in swine at slaughterhouse in Italy. Vet Microbiol. 2011; 149:330–338. PMID: 21216541.3. Colson P, Romanet P, Moal V, Borentain P, Purgus R, Benezech A, et al. Autochthonous infections with hepatitis E virus genotype 4, France. Emerg Infect Dis. 2012; 18:1361–1364. PMID: 22840196.4. Kumar A, Beniwal M, Kar P, Sharma JB, Murthy NS. Hepatitis E in pregnancy. Int J Gynaecol Obstet. 2004; 85:240–244. PMID: 15145258.5. Smith DB, Purdy MA, Simmonds P. Genetic variability and the classification of hepatitis E virus. J Virol. 2013; 87:4161–4169. PMID: 23388713.6. De Silva AN, Muddu AK, Iredale JP, Sheron N, Khakoo SI, Pelosi E. Unexpectedly high incidence of indigenous acute hepatitis E within South Hampshire: time for routine testing? J Med Virol. 2008; 80:283–288. PMID: 18098134.7. Pavio N, Meng XJ, Renou C. Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res. 2010; 41:46. PMID: 20359452.8. Berto A, Martelli F, Grierson S, Banks M. Hepatitis E virus in pork food chain, United Kingdom, 2009–2010. Emerg Infect Dis. 2012; 18:1358–1360. PMID: 22840183.9. Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008; 48:494–503. PMID: 18192058.10. van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, et al. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg Infect Dis. 2001; 7:970–976. PMID: 11747723.11. Takahashi K, Kitajima N, Abe N, Mishiro S. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology. 2004; 330:501–505. PMID: 15567444.12. Meng XJ. Recent advances in Hepatitis E virus. J Viral Hepat. 2010; 17:153–161. PMID: 20040046.13. Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002; 40:1326–1332. PMID: 11923352.14. Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Nat Acad Sci USA. 1997; 94:9860–9865. PMID: 9275216.15. Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011; 161:23–30. PMID: 21316404.16. Pavio N, Merbah T, Thébault A. Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg Infect Dis. 2014; 20:1925–1927. PMID: 25340373.17. Mizuo H, Yazaki Y, Sugawara K, Tsuda F, Takahashi M, Nishizawa T, et al. Possible risk factors for the transmission of hepatitis E virus and for the severe form of hepatitis E acquired locally in Hokkaido, Japan. J Med Virol. 2005; 76:341–349. PMID: 15902701.18. Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991; 185:120–131. PMID: 1926770.19. Meng J, Dai X, Chang JC, Lopareva E, Pillot J, Fields HA, et al. Identification and characterization of the neutralization epitope(s) of the hepatitis E virus. Virology. 2001; 288:203–211. PMID: 11601892.20. Jiménez de Oya N, Galindo I, Gironés O, Duizer E, Escribano JM, Saiz JC. Serological immunoassay for detection of hepatitis E virus on the basis of genotype 3 open reading frame 2 recombinant proteins produced in Trichoplusia ni larvae. J Clin Microbiol. 2009; 47:3276–3282. PMID: 19656986.21. Baechlein C, Meemken D, Pezzoni G, Engemann C, Grummer B. Expression of a truncated hepatitis E virus capsid protein in the protozoan organism Leishmania tarentolae and its application in a serological assay. J Virol Methods. 2013; 193:238–243. PMID: 23747546.22. Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Nat Acad Sci USA. 2009; 106:12986–12991. PMID: 19620712.23. Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med. 2012; 367:1237–1244. PMID: 23013075.24. Bendall R, Ellis V, Ijaz S, Ali R, Dalton H. A comparison of two commercially available anti-HEV IgG kits and a re-evaluation of anti-HEV IgG seroprevalence data in developed countries. J Med Virol. 2010; 82:799–805. PMID: 20336757.25. Mett V, Farrance CE, Green BJ, Yusibov V. Plants as biofactories. Biologicals. 2008; 36:354–358. PMID: 18938088.26. Lomonossoff GP, D'Aoust MA. Plant-produced biopharmaceuticals: A case of technical developments driving clinical deployment. Science. 2016; 353:1237–1240. PMID: 27634524.27. Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009; 7:682–693. PMID: 19627561.28. Love AJ, Chapman SN, Matic S, Noris E, Lomonossoff GP, Taliansky M. In planta production of a candidate vaccine against bovine papillomavirus type 1. Planta. 2012; 236:1305–1313. PMID: 22718313.29. Taherkhani R, Makvandi M, Farshadpour F. Development of enzyme-linked immunosorbent assays using 2 truncated ORF2 proteins for detection of IgG antibodies against hepatitis E virus. Ann Lab Med. 2014; 34:118–126. PMID: 24624347.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development of Enzyme-Linked Immunosorbent Assays Using 2 Truncated ORF2 Proteins for Detection of IgG Antibodies Against Hepatitis E Virus
- Current Status of Hepatitis E Virus Infection in Korea
- Development of a novel enzyme-linked immunosorbent assay to detect anti-IgG against swine hepatitis E virus
- Replication of Recombinant Flock House Virus RNA Encapsidated by Turnip Yellow Mosaic Virus Coat Proteins in Nicotiana benthamiana
- Detection and Genetic Characterization of Isolates of Hepatitis E Virus from Pigs and Human in Chungnam Region of Korea