Ann Lab Med.
2016 Sep;36(5):475-480. 10.3343/alm.2016.36.5.475.
Frequency and Clinical Characteristics of Intrachromosomal Amplification of Chromosome 21 in Korean Childhood B-lineage Acute Lymphoblastic Leukemia
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea. LEE.ST@yuhs.ac ugine01@naver.com
- 2Department of Pediatrics, Yonsei Cancer Research Center, Yonsei University College of Medicine, Seoul, Korea.
- 3Department of Laboratory Medicine, Hallym University College of Medicine, Kangnam Sacred Heart Hospital, Seoul, Korea. LEE.ST@yuhs.ac ugine01@naver.com
- KMID: 2373579
- DOI: http://doi.org/10.3343/alm.2016.36.5.475
Abstract
- BACKGROUND
Intrachromosomal amplification of chromosome 21 (iAMP21) is known to be associated with poor prognosis in B-cell ALL (B-ALL). To determine the frequency and clinical characteristics of iAMP21 in Korean B-ALL patients, we performed FISH and multiplex ligation-dependent probe amplification (MLPA) analyses.
METHODS
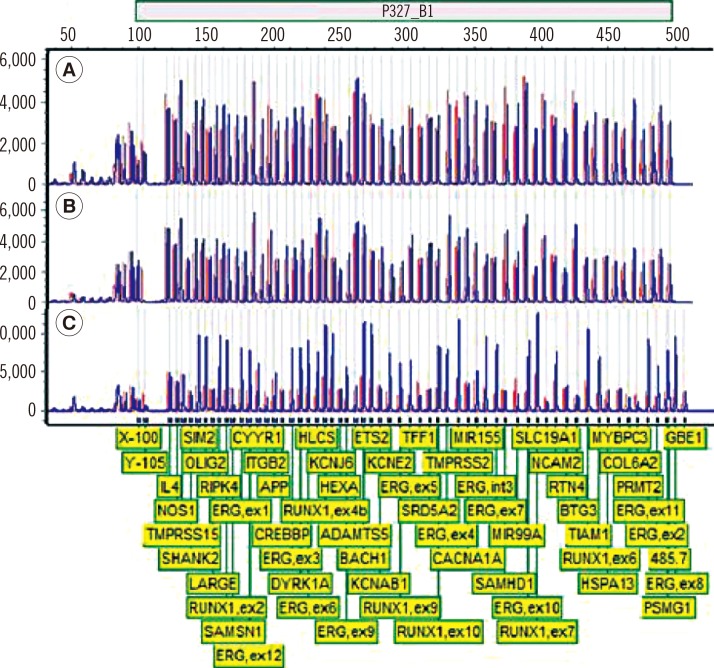
A total of 102 childhood B-ALL patients were screened with ETV6-RUNX1 FISH probes (Abbott Molecular, USA). The presence of an iAMP21 was confirmed by using MLPA P327 iAMP21-ERG probemix (MRC Holland, The Netherlands).
RESULTS
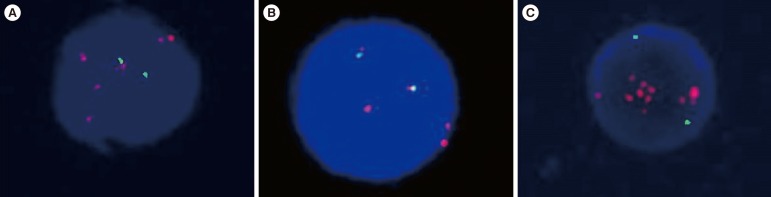
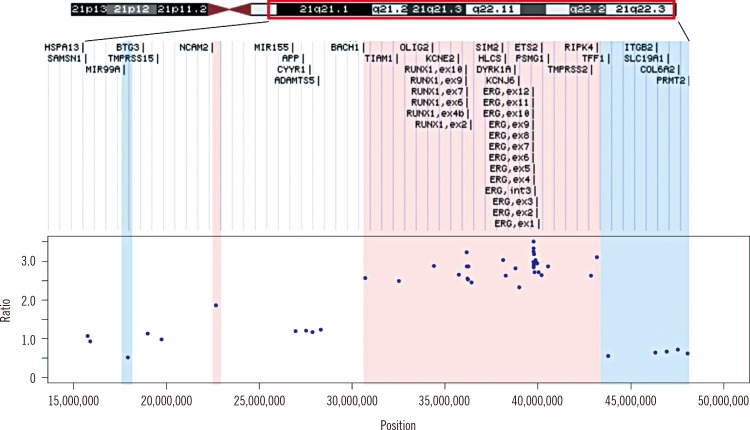
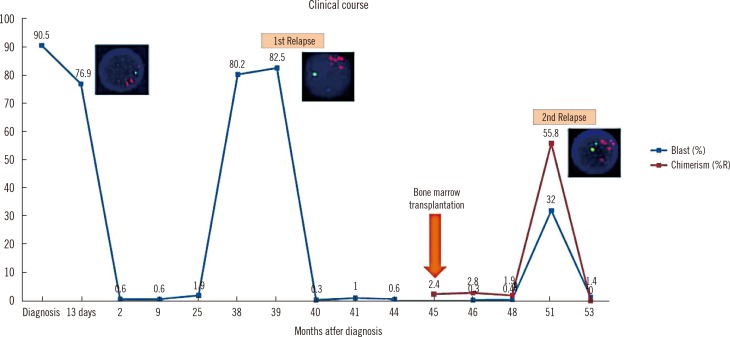
iAMP21 was detected in one of the screened B-ALL patients (1/102 patients, 1.0%) who presented the ALL immunophenotype and complex karyotype at initial diagnosis. The patient relapsed twice after bone marrow transplantation. MLPA showed 12.5-Mb and 4.28-Mb regions of amplification and deletion, respectively.
CONCLUSIONS
The frequency of iAMP21 is considerable in Korean pediatric patients. Our report suggests that iAMP21 in childhood B-ALL has very unfavorable impact on patient's prognosis. Additional methods such as MLPA analysis is essential to rule out patients with equivocal interphase FISH results.
MeSH Terms
-
Adolescent
Asian Continental Ancestry Group/*genetics
B-Lymphocytes/*metabolism
Child
Child, Preschool
*Chromosomes, Human, Pair 21
Core Binding Factor Alpha 2 Subunit/genetics
DNA Probes/metabolism
Female
Humans
Immunophenotyping
In Situ Hybridization, Fluorescence
Infant
Infant, Newborn
Male
Multiplex Polymerase Chain Reaction
Precursor Cell Lymphoblastic Leukemia-Lymphoma/*diagnosis/genetics
Proto-Oncogene Proteins c-ets/genetics
Repressor Proteins/genetics
Republic of Korea
Translocation, Genetic
Young Adult
Core Binding Factor Alpha 2 Subunit
DNA Probes
Proto-Oncogene Proteins c-ets
Repressor Proteins
Figure
Cited by 1 articles
-
Recent advances in the treatment of pediatric acute leukemia
Hyery Kim
J Korean Med Assoc. 2016;59(9):690-697. doi: 10.5124/jkma.2016.59.9.690.
Reference
-
1. Pui C-H. Childhood leukemias. 3rd ed. Cambridge: Cambridge University Press;2012. p. 880 S.2. Harewood L, Robinson H, Harris R, Al-Obaidi MJ, Jalali GR, Martineau M, et al. Amplification of AML1 on a duplicated chromosome 21 in acute lymphoblastic leukemia: a study of 20 cases. Leukemia. 2003; 17:547–553. PMID: 12646943.3. Harrison CJ, Moorman AV, Barber KE, Broadfield ZJ, Cheung KL, Harris RL, et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group Study. Br J Haematol. 2005; 129:520–530. PMID: 15877734.4. Harrison CJ, Haas O, Harbott J, Biondi A, Stanulla M, Trka J, et al. Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfürt-Münster study group. Br J Haematol. 2010; 151:132–142. PMID: 20701601.5. Reichard KK, Kang H, Robinett S. Pediatric B-lymphoblastic leukemia with RUNX1 amplification: clinicopathologic study of eight cases. Mod Pathol. 2011; 24:1606–1611. PMID: 21822204.6. Moorman AV, Richards SM, Robinson HM, Strefford JC, Gibson BE, Kinsey SE, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21). Blood. 2007; 109:2327–2330. PMID: 17095619.7. Harrison CJ. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood. 2015; 125:1383–1386. PMID: 25608562.8. Strefford JC, van Delft FW, Robinson HM, Worley H, Yiannikouris O, Selzer R, et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc Natl Acad Sci U S A. 2006; 103:8167–8172. PMID: 16702559.9. Harrison CJ, Moorman AV, Schwab C, Carroll AJ, Raetz EA, Devidas M, et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia. 2014; 28:1015–1021. PMID: 24166298.10. Rand V, Parker H, Russell LJ, Schwab C, Ensor H, Irving J, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011; 117:6848–6855. PMID: 21527530.11. Robinson HM, Harrison CJ, Moorman AV, Chudoba I, Strefford JC. Intrachromosomal amplification of chromosome 21 (iAMP21) may arise from a breakage-fusion-bridge cycle. Genes Chromosomes Cancer. 2007; 46:318–326. PMID: 17243167.12. Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009; 144:147–156. PMID: 19006567.13. Dirse V, Bertasiute A, Gineikiene E, Zvirblis T, Dambrauskiene R, Gerbutavicius R, et al. A population-based single nucleotide polymorphism array analysis of genomic aberrations in younger adult acute lymphoblastic leukemia patients. Genes Chromosomes Cancer. 2015; 54:326–333. PMID: 25706938.14. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002; 30:e57. PMID: 12060695.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Intrachromosomal amplification of chromosome 21 in Korean pediatric patients with B-cell precursor acute lymphoblastic leukemia in a single institution
- TEL/AML1 Fusion Transcripts in Childhood B-Lineage Acute Lymphoblastic Leukemia
- Lineage Switch at Relapse of Childhood Acute Leukemia: A Report of Four Cases
- Two Cases of Chronic Myeloid Leukemia in Lymphoid Blast Phase Presented as Philadelphia-Positive Acute Lymphoblastic Leukemia
- Clinical Significance of CD34 Antigen Positivity in Childhood Acute Lymphoblastic Leukemia