Ann Lab Med.
2016 Sep;36(5):441-449. 10.3343/alm.2016.36.5.441.
Analysis of the Vaginal Microbiome by Next-Generation Sequencing and Evaluation of its Performance as a Clinical Diagnostic Tool in Vaginitis
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul Medical Center, Seoul, Korea.
- 2Department of Laboratory Medicine, College of Medicine, Seoul National University, Seoul, Korea.
- 3Department of Laboratory Medicine, Sheikh Khalifa Specialty Hospital, Ras Al Khaimah, United Arab Emirates. mwseong@snu.ac.kr
- 4Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea.
- 5Department of Obstetrics & Gynecology, Seoul National University Hospital Healthcare System Gangnam Center, Seoul, Korea.
- 6Department of Obstetrics & Gynecology, Konkuk University Medical Center, Seoul, Korea.
- KMID: 2373574
- DOI: http://doi.org/10.3343/alm.2016.36.5.441
Abstract
- BACKGROUND
Next-generation sequencing (NGS) can detect many more microorganisms of a microbiome than traditional methods. This study aimed to analyze the vaginal microbiomes of Korean women by using NGS that included bacteria and other microorganisms. The NGS results were compared with the results of other assays, and NGS was evaluated for its feasibility for predicting vaginitis.
METHODS
In total, 89 vaginal swab specimens were collected. Microscopic examinations of Gram staining and microbiological cultures were conducted on 67 specimens. NGS was performed with GS junior system on all of the vaginal specimens for the 16S rRNA, internal transcribed spacer (ITS), and Tvk genes to detect bacteria, fungi, and Trichomonas vaginalis. In addition, DNA probe assays of the Candida spp., Gardnerella vaginalis, and Trichomonas vaginalis were performed. Various predictors of diversity that were obtained from the NGS data were analyzed to predict vaginitis.
RESULTS
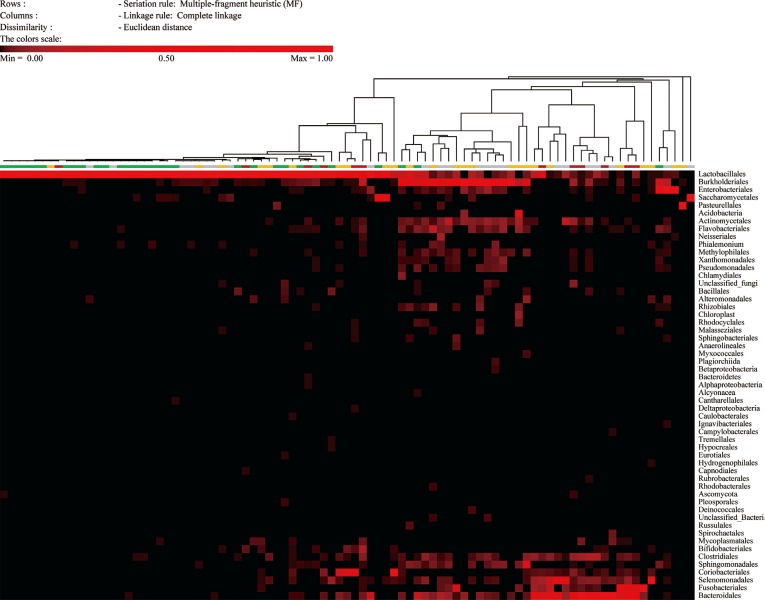
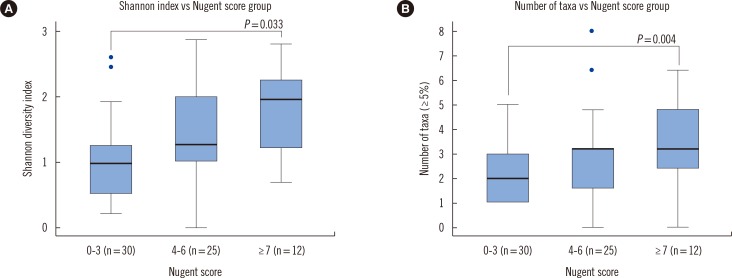
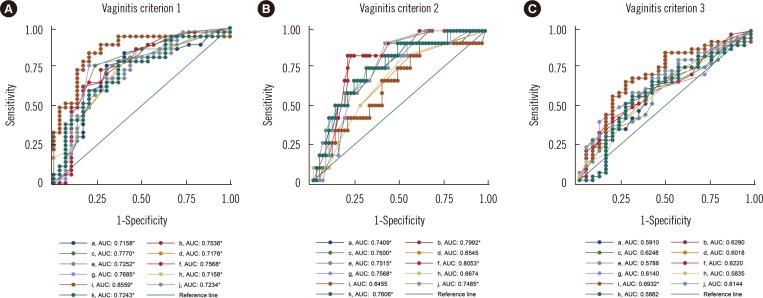
ITS sequences were obtained in most of the specimens (56.2%). The compositions of the intermediate and vaginitis Nugent score groups were similar to each other but differed from the composition of the normal score group. The fraction of the Lactobacillus spp. showed the highest area under the curve value (0.8559) in ROC curve analysis. The NGS and DNA probe assay results showed good agreement (range, 86.2-89.7%).
CONCLUSIONS
Fungi as well as bacteria should be considered for the investigation of vaginal microbiome. The intermediate and vaginitis Nugent score groups were indistinguishable in NGS. NGS is a promising diagnostic tool of the vaginal microbiome and vaginitis, although some problems need to be resolved.
Keyword
MeSH Terms
-
Area Under Curve
Bacteria/*genetics/isolation & purification
Bacterial Proteins/genetics
Candida/*genetics/isolation & purification
Female
Fungal Proteins/genetics
Gardnerella vaginalis/genetics/isolation & purification
High-Throughput Nucleotide Sequencing
Humans
*Microbiota
RNA, Ribosomal, 16S/chemistry/genetics/metabolism
ROC Curve
Sequence Analysis, DNA
Trichomonas vaginalis/genetics/isolation & purification
Vagina/*microbiology
Vaginitis/*diagnosis/microbiology
Bacterial Proteins
Fungal Proteins
RNA, Ribosomal, 16S
Figure
Reference
-
1. Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, Gloor GB, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010; 5:e12078. PMID: 20711427.2. Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, et al. Molecular definition of vaginal microbiota in East African commercial sex workers. Appl Environ Microbiol. 2011; 77:4066–4074. PMID: 21531840.3. Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010; 11:488. PMID: 20819230.4. Liu MB, Xu SR, He Y, Deng GH, Sheng HF, Huang XM, et al. Diverse vaginal microbiomes in reproductive-age women with vulvovaginal candidiasis. PLoS One. 2013; 8:e79812. PMID: 24265786.5. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011; 108(Suppl 1):4680–4687. PMID: 20534435.6. Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991; 29:297–301. PMID: 1706728.7. Rodrigues FS, Peixoto S, Adami F, Alves Bda C, Gehrke Fde S, Azzalis LA, et al. Proposal of a new cutoff for Nugent criteria in the diagnosis of bacterial vaginosis. J Microbiol Methods. 2015; 115:144–146. PMID: 25957514.8. Jumpstart Consortium Human Microbiome Project Data Generation Working Group. 16S 454 Sequencing Protocol. Updated on Oct 27, 2010. http://www.hmpdacc.org/doc/16S_Sequencing_SOP_4.2.2.pdf.9. CLSI. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; Approved guideline. CLSI document MM18-A. Wayne, PA: Clinical and Laboratory Standards Institute;2008.10. Lawing LF, Hedges SR, Schwebke JR. Detection of trichomonosis in vaginal and urine specimens from women by culture and PCR. J Clin Microbiol. 2000; 38:3585–3588. PMID: 11015368.11. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. PMID: 20709691.12. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011; 27:2194–2200. PMID: 21700674.13. Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011; 21:494–504. PMID: 21212162.14. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007; 73:5261–5267. PMID: 17586664.15. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215:403–410. PMID: 2231712.16. Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005; 21:1280–1281. PMID: 15546938.17. Shannon CE. A mathematical theory of communication. Bell System Tech J. 1948; 27:379–423.18. Martin DH, Zozaya M, Lillis R, Miller J, Ferris MJ. The microbiota of the human genitourinary tract: trying to see the forest through the trees. Trans Am Clin Climatol Assoc. 2012; 123:242–256. PMID: 23303991.19. Smidt I, Kiiker R, Oopkaup H, Lapp E, Rööp T, Truusalu K, et al. Comparison of detection methods for vaginal lactobacilli. Benef Microbes. 2015; 6:747–751. PMID: 25869280.20. Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One. 2013; 8:e65226. PMID: 23734239.21. Toma I, Siegel MO, Keiser J, Yakovleva A, Kim A, Davenport L, et al. Single-molecule long-read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol. 2014; 52:3913–3921. PMID: 25143582.22. Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2012; 13(Suppl 8):S17. PMID: 23282177.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Vaginal Stenosis due to Chemical Vaginitis after Using Jamu Sticks: A Case Report
- Microbiome Analysis Using Next-Generation Sequencing in Urinary Tract Infections
- Vaginal health in women and the possibility of predicting preterm birth through microbiome analysis
- Diagnostic Values of pH of Vaginal Discharge, Amine Test, and Microscopic Examination of Infectious Vaginitis
- A Case of Neonatal Trichomonas Vaginitis