Obstet Gynecol Sci.
2015 Jul;58(4):261-267. 10.5468/ogs.2015.58.4.261.
Explore the dynamic alternation of gene PLAC4 mRNA expression levels in maternal plasma in second trimester for nonivasive detection of trisomy 21
- Affiliations
-
- 1Department of Prenatal Diagnosis Center, Wuxi Maternal and Child Health Hospital Affiliated Nanjing Medical University, Wuxi, China. lilylan5930@sina.com
- 2State Key Laboratory of Genetic Engineering, Fu Dan University, Shanghai, China.
- KMID: 2314045
- DOI: http://doi.org/10.5468/ogs.2015.58.4.261
Abstract
OBJECTIVE
Noninvasive prenatal detection of trisomy 21 (T21) has been achieved by measuring the ratio of two alleles of a single nucleotide polymorphism in circulating placenta specific 4 (PLAC4) mRNA in maternal plasma with a few assays in recent years. Our research is to explore the variations of PLAC4 mRNA expression level in maternal plasma with normal pregnancies in second trimester, which can provide pregnant women deeper insights with suitable detection period for the non-invasive prenatal detection of T21.
METHODS
We measured a serial plasma PLAC4 mRNA concentrations weekly from the same 25 singleton normal pregnant women. We recruited maternal plasma samples from 45 singleton pregnant women, comprising of 25 euploid pregnancies (control group; range, 17 to 21 weeks) and 20 T21 pregnancies (T21 group; range, 19 to 24 weeks). With the application of reverse transcription polymerase chain reaction, we achieved an insight of PLAC4 mRNA expression levels in maternal plasma during second trimester with euploid pregnancies.
RESULTS
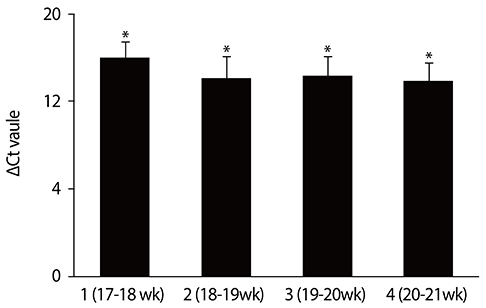
Among the control group, the levels of PLAC4 mRNA expression in the gestation of 17 to 18 weeks were significantly less than those in the gestation of 18 to 21 weeks (P<0.05). The average PLAC4 mRNA concentration of the normal pregnant women was not higher than that of the T21 group (P>0.05).
CONCLUSION
The PLAC4 mRNA showed a higher level of expression in the gestation of 18 to 21 weeks with an euploid pregnancy of pregnant women. We also found that there was no significant difference in plasma PLAC4 mRNA concentration between the normal and the T21 pregnancies in second trimester.
Keyword
MeSH Terms
Figure
Reference
-
1. Alfirevic Z, Sundberg K, Brigham S. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst Rev. 2003; CD003252.2. Malone FD, Canick JA, Ball RH, Nyberg DA, Comstock CH, Bukowski R, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med. 2005; 353:2001–2011.3. Wright D, Syngelaki A, Bradbury I, Akolekar R, Nicolaides KH. First-trimester screening for trisomies 21, 18 and 13 by ultrasound and biochemical testing. Fetal Diagn Ther. 2014; 35:118–126.4. Breathnach FM, Malone FD. Screening for aneuploidy in first and second trimesters: is there an optimal paradigm? Curr Opin Obstet Gynecol. 2007; 19:176–182.5. Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010; 28:827–838.6. Nicolaides KH, Syngelaki A, Poon LC, Gil MM, Wright D. First-trimester contingent screening for trisomies 21, 18 and 13 by biomarkers and maternal blood cell-free DNA testing. Fetal Diagn Ther. 2014; 35:185–192.7. Tsui NB, Akolekar R, Chiu RW, Chow KC, Leung TY, Lau TK, et al. Synergy of total PLAC4 RNA concentration and measurement of the RNA single-nucleotide polymorphism allelic ratio for the noninvasive prenatal detection of trisomy 21. Clin Chem. 2010; 56:73–81.8. Yang L, Sun H, Chen D, Lu M, Wang J, Xu F, et al. Application of multiplex SNaPshot assay in measurement of PLAC4 RNA-SNP allelic ratio for noninvasive prenatal detection of trisomy 21. Prenat Diagn. 2014; 34:139–144.9. Lo YM, Tsui NB, Chiu RW, Lau TK, Leung TN, Heung MM, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat Med. 2007; 13:218–223.10. Liu HY, Yang LL, Li F, Yang HJ, Liu SM, Zhang JB. Detection and clinical significance of fetal specific mRNA from peripheral maternal blood. Zhonghua Fu Chan Ke Za Zhi. 2011; 46:655–657.11. Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002; 48:1647–1653.12. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010; 50:298–301.13. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008; 105:10513–10518.14. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408.15. Li QF, Jiang MY, Yu HX, SW Xin, MH Gu, QQ Liu. Selection of internal reference genes for quantitative RT-PCR analysis of total RNA from endosperm of rice (Oryza sativa L.). J Yangzhou Univ. 2008; 29:61–66.16. Scharlaken B, de Graaf DC, Goossens K, Brunain M, Peelman LJ, Jacobs FJ. Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. J Insect Sci. 2008; 8:33.17. Tsui NB, Chim SS, Chiu RW, Lau TK, Ng EK, Leung TN, et al. Systematic micro-array based identification of placental mRNA in maternal plasma: towards non-invasive prenatal gene expression profiling. J Med Genet. 2004; 41:461–467.18. Kido S, Sakuragi N, Bronner MP, Sayegh R, Berger R, Patterson D, et al. D21S418E identifies a cAMP-regulated gene located on chromosome 21q22.3 that is expressed in placental syncytiotrophoblast and choriocarcinoma cells. Genomics. 1993; 17:256–259.19. Volk M, Maver A, Lovrecic L, Juvan P, Peterlin B. Expression signature as a biomarker for prenatal diagnosis of trisomy 21. PLoS One. 2013; 8:e74184.20. Middlebrooks CD, Mukhopadhyay N, Tinker SW, Allen EG, Bean LJ, Begum F, et al. Evidence for dysregulation of genome-wide recombination in oocytes with nondisjoined chromosomes 21. Hum Mol Genet. 2014; 23:408–417.21. Mao R, Wang X, Spitznagel EL Jr, Frelin LP, Ting JC, Ding H, et al. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 2005; 6:R107.22. Li CM, Guo M, Salas M, Schupf N, Silverman W, Zigman WB, et al. Cell type-specific over-expression of chromosome 21 genes in fibroblasts and fetal hearts with trisomy 21. BMC Med Genet. 2006; 7:24.23. Conti A, Fabbrini F, D'Agostino P, Negri R, Greco D, Genesio R, et al. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genomics. 2007; 8:268.24. Kirschner MB, van Zandwijk N, Reid G. Cell-free microRNAs: potential biomarkers in need of standardized reporting. Front Genet. 2013; 4:56.25. Altug-Teber O, Bonin M, Walter M, Mau-Holzmann UA, Dufke A, Stappert H, et al. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007; 119:171–184.26. FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum Mol Genet. 2002; 11:3249–3256.27. Rozovski U, Jonish-Grossman A, Bar-Shira A, Ochshorn Y, Goldstein M, Yaron Y. Genome-wide expression analysis of cultured trophoblast with trisomy 21 karyotype. Hum Reprod. 2007; 22:2538–2545.28. Chou CY, Liu LY, Chen CY, Tsai CH, Hwa HL, Chang LY, et al. Gene expression variation increase in trisomy 21 tissues. Mamm Genome. 2008; 19:398–405.29. Slonim DK, Koide K, Johnson KL, Tantravahi U, Cowan JM, Jarrah Z, et al. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in Down syndrome fetuses. Proc Natl Acad Sci U S A. 2009; 106:9425–9429.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- First trimester screening for trisomy 18 by a combination of nuchal translucency thickness and epigenetic marker level
- Screening for chromosomal abnormalities using combined test in the first trimester of pregnancy
- The predictive value of abnormal ultrasonographic finding for fetal trisomy in the second trimester
- The effect of a vanishing twin on first- and second-trimester maternal serum markers and ultrasound screening for aneuploidy
- Detection of trisomy 9 mosaicism in the second trimester screening by abnormal level of biochemical markers