J Breast Cancer.
2014 Sep;17(3):207-218. 10.4048/jbc.2014.17.3.207.
The Role and Regulatory Mechanism of 14-3-3 Sigma in Human Breast Cancer
- Affiliations
-
- 1Department of Surgery, Cheil General Hospital & Women's Health Care Center, Catholic Kwandong University College of Medicine, Seoul, Korea.
- 2Department of Pathology, CHA Gangnam Medical Center, CHA University, Seoul, Korea.
- 3Department of Surgery, Yonsei University College of Medicine, Seoul, Korea.
- 4Department of Anatomy, Yonsei University College of Medicine, Seoul, Korea.
- 5Department of Pathology, Yonsei University College of Medicine, Seoul, Korea. jungwh96@yuhs.ac
- KMID: 2286347
- DOI: http://doi.org/10.4048/jbc.2014.17.3.207
Abstract
- PURPOSE
14-3-3 sigma (sigma) is considered to be an important tumor suppressor and decreased expression of the same has been reported in many malignant tumors by hypermethylation at its promoter or ubiquitin-mediated proteolysis by estrogen-responsive ring finger protein (Efp). In this study, we investigated the significance of 14-3-3 sigma expression in human breast cancer and its regulatory mechanism.
METHODS
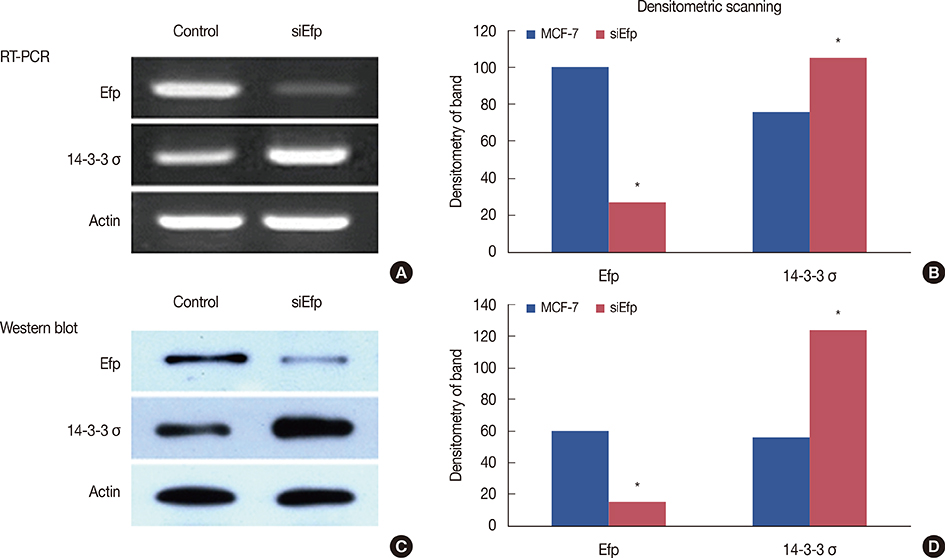
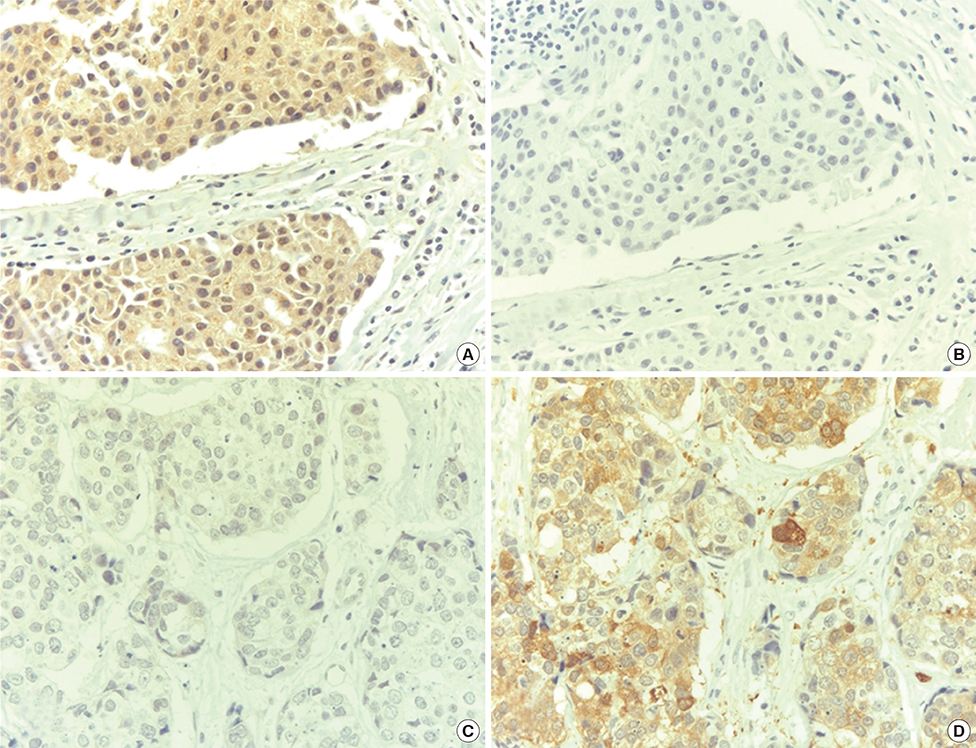
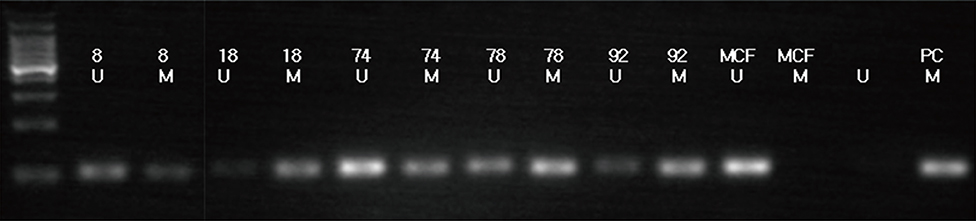
Efp was silenced using small interfering RNA (siRNA) in the MCF-7 breast cancer cell line in order to examine its influence on the level of 14-3-3 sigma protein. The methylation status of the 14-3-3 sigma promoter was also evaluated by methylation-specific polymerase chain reaction (PCR). The expression of Efp and 14-3-3 sigma in 220 human breast carcinoma tissues was assessed by immunohistochemistry. Other clinicopathological parameters were also evaluated.
RESULTS
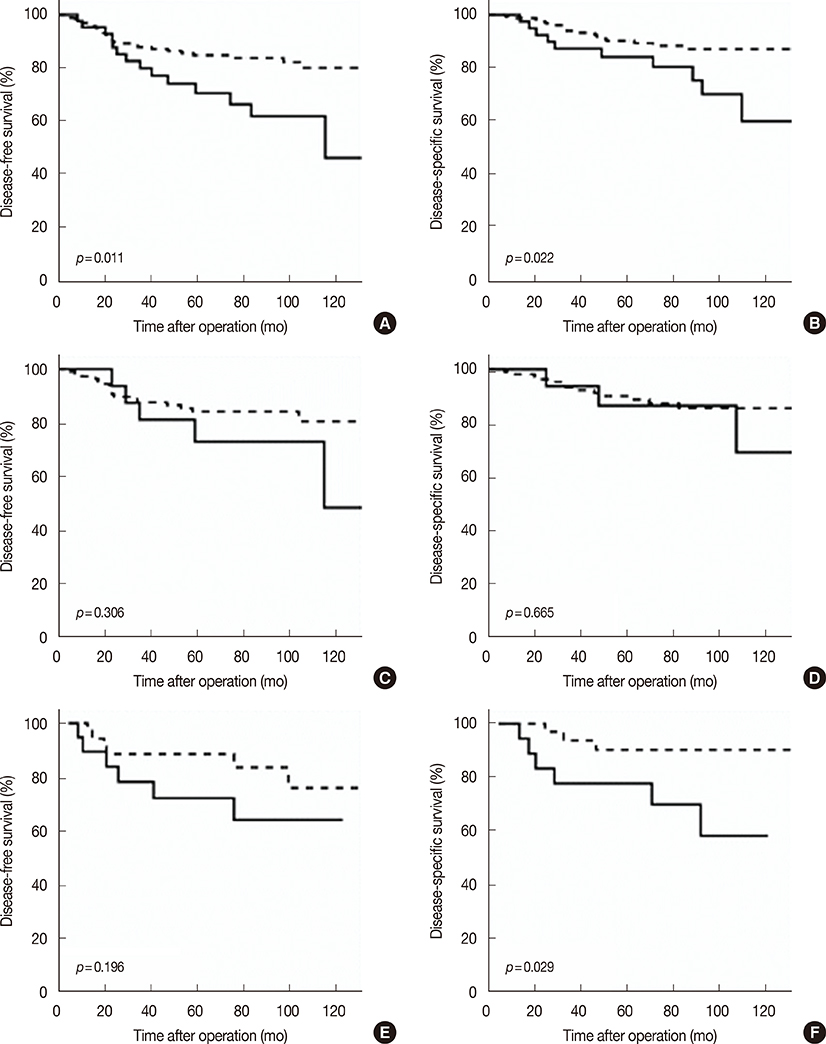
Silencing Efp in the MCF-7 breast cancer cell line resulted in increased expression of 14-3-3 sigma. The Efp-positive human breast cancers were more frequently 14-3-3 sigma-negative (60.5% vs. 39.5%). Hypermethylation of 14-3-3 sigma was common (64.9%) and had an inverse association with 14-3-3 sigma positivity (p=0.072). Positive 14-3-3 sigma expression was significantly correlated with poor prognosis: disease-free survival (p=0.008) and disease-specific survival (p=0.009).
CONCLUSION
Our data suggests that in human breast cancer, the regulation of 14-3-3 sigma may involve two mechanisms: ubiquitin-mediated proteolysis by Efp and downregulation by hypermethylation. However, the inactivation of 14-3-3 sigma is probably achieved mainly by hypermethylation. Interestingly, 14-3-3 sigma turned out to be a very significant poor prognostic indicator, which is in contrast to its previously known function as a tumor suppressor, suggesting a different role of 14-3-3 sigma in breast cancer.
MeSH Terms
Figure
Reference
-
1. Ko SS. Korean Breast Cancer Society. Chronological changing patterns of clinical characteristics of Korean breast cancer patients during 10 years (1996-2006) using nationwide breast cancer registration on-line program: biannual update. J Surg Oncol. 2008; 98:318–323.
Article2. Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997; 1:3–11.3. Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999; 401:616–620.
Article4. Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol. 2003; 23:7096–7107.
Article5. Gasco M, Bell AK, Heath V, Sullivan A, Smith P, Hiller L, et al. Epigenetic inactivation of 14-3-3 sigma in oral carcinoma: association with p16(INK4a) silencing and human papillomavirus negativity. Cancer Res. 2002; 62:2072–2076.6. Osada H, Tatematsu Y, Yatabe Y, Nakagawa T, Konishi H, Harano T, et al. Frequent and histological type-specific inactivation of 14-3-3sigma in human lung cancers. Oncogene. 2002; 21:2418–2424.
Article7. Lodygin D, Diebold J, Hermeking H. Prostate cancer is characterized by epigenetic silencing of 14-3-3sigma expression. Oncogene. 2004; 23:9034–9041.
Article8. Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 sigma gene is associated with 5' CpG island hypermethylation in human cancers. Cancer Res. 2000; 60:4353–4357.9. Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, et al. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000; 97:6049–6054.
Article10. Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S. Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene. 2001; 20:3348–3353.
Article11. Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002; 417:871–875.
Article12. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991; 19:403–410.
Article13. Pertschuk LP, Feldman JG, Kim YD, Braithwaite L, Schneider F, Braverman AS, et al. Estrogen receptor immunocytochemistry in paraffin embedded tissues with ER1D5 predicts breast cancer endocrine response more accurately than H222Sp gamma in frozen sections or cytosol-based ligand-binding assays. Cancer. 1996; 77:2514–2519.
Article14. De Potter CR, Quatacker J, Maertens G, Van Daele S, Pauwels C, Verhofstede C, et al. The subcellular localization of the neu protein in human normal and neoplastic cells. Int J Cancer. 1989; 44:969–974.
Article15. Styles JM, Harrison S, Gusterson BA, Dean CJ. Rat monoclonal antibodies to the external domain of the product of the C-erbB-2 proto-oncogene. Int J Cancer. 1990; 45:320–324.
Article16. Marchetti A, Buttitta F, Pellegrini S, Campani D, Diella F, Cecchetti D, et al. p53 mutations and histological type of invasive breast carcinoma. Cancer Res. 1993; 53:4665–4669.17. Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci U S A. 1992; 89:7262–7266.
Article18. Kaneuchi M, Sasaki M, Tanaka Y, Shiina H, Verma M, Ebina Y, et al. Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer. Biochem Biophys Res Commun. 2004; 316:1156–1162.
Article19. Lehmann U, Länger F, Feist H, Glöckner S, Hasemeier B, Kreipe H. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol. 2002; 160:605–612.
Article20. Simooka H, Oyama T, Sano T, Horiguchi J, Nakajima T. Immunohistochemical analysis of 14-3-3 sigma and related proteins in hyperplastic and neoplastic breast lesions, with particular reference to early carcinogenesis. Pathol Int. 2004; 54:595–602.
Article21. Horie K, Urano T, Ikeda K, Inoue S. Estrogen-responsive RING finger protein controls breast cancer growth. J Steroid Biochem Mol Biol. 2003; 85:101–104.
Article22. Suzuki T, Urano T, Tsukui T, Horie-Inoue K, Moriya T, Ishida T, et al. Estrogen-responsive finger protein as a new potential biomarker for breast cancer. Clin Cancer Res. 2005; 11:6148–6154.
Article23. Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T, et al. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc Natl Acad Sci U S A. 1993; 90:11117–11121.
Article24. Ikeda K, Orimo A, Higashi Y, Muramatsu M, Inoue S. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000; 472:9–13.
Article25. Thomson SD, Ali S, Pickles L, Taylor J, Pace PE, Lymboura M, et al. Analysis of estrogen-responsive finger protein expression in benign and malignant human breast. Int J Cancer. 2001; 91:152–158.
Article26. Ikeda K, Inoue S, Orimo A, Sano M, Watanabe T, Tsutsumi K, et al. Multiple regulatory elements and binding proteins of the 5\'-flanking region of the human estrogen-responsive finger protein (efp) gene. Biochem Biophys Res Commun. 1997; 236:765–771.
Article27. Perathoner A, Pirkebner D, Brandacher G, Spizzo G, Stadlmann S, Obrist P, et al. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin Cancer Res. 2005; 11:3274–3279.
Article28. Nakayama H, Sano T, Motegi A, Oyama T, Nakajima T. Increasing 14-3-3 sigma expression with declining estrogen receptor alpha and estrogen-responsive finger protein expression defines malignant progression of endometrial carcinoma. Pathol Int. 2005; 55:707–715.
Article29. Voutsadakis IA. The ubiquitin-proteasome system in colorectal cancer. Biochim Biophys Acta. 2008; 1782:800–808.
Article30. Dawson SP. Hepatocellular carcinoma and the ubiquitin-proteasome system. Biochim Biophys Acta. 2008; 1782:775–784.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of the 14-3-3 sigma Protein and Methylation Status of the 14-3-3 sigma gene in Biliary Neoplasms
- Characterization of Chromatin Structure-associated Histone Modifications in Breast Cancer Cells
- The Role of Preoperative Breast MRI in Patients With Early-Stage Breast Cancer
- The Highligts of 28th Annual Meeting of San Antonio Breast Cancer Symposium
- Diagnosis and Treatment of HER2-Positive Breast Cancer