Blood Res.
2013 Sep;48(3):206-210. 10.5045/br.2013.48.3.206.
Sequence variation data of F8 and F9 genes in functionally validated control individuals: implications on the molecular diagnosis of hemophilia
- Affiliations
-
- 1Department of Laboratory Medicine & Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. heejinkim@skku.edu
- 2Samsung Biomedical Research Institute, Samsung Medical Center, Seoul, Korea.
- KMID: 2270707
- DOI: http://doi.org/10.5045/br.2013.48.3.206
Abstract
- BACKGROUND
The F8 and F9 genes encode for coagulation factor VIII (FVIII) and FIX, respectively, and mutations in these genes are the genetic basis of hemophilia A/B. To determine whether a sequence variation in F8/F9 is a disease-causing mutation, frequency data from a control population is needed. This study aimed to obtain data on sequence variation in F8/F9 in a set of functionally validated control chromosomes of Korean descent.
METHODS
We re-sequenced F8 and F9 from DNA samples of 100 Korean male control individuals with normal PT, aPTT, and FVIII activity. PCR and direct sequencing analyses were performed using primer pairs to cover all coding regions and the flanking intronic sequences.
RESULTS
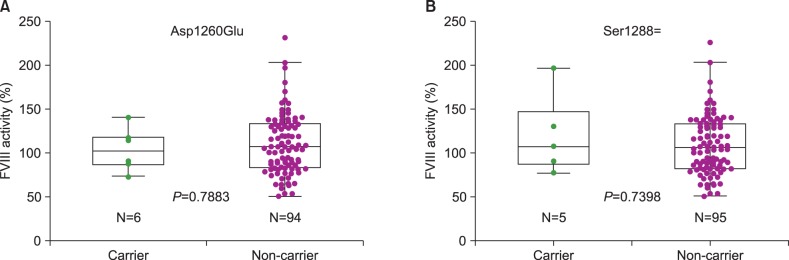
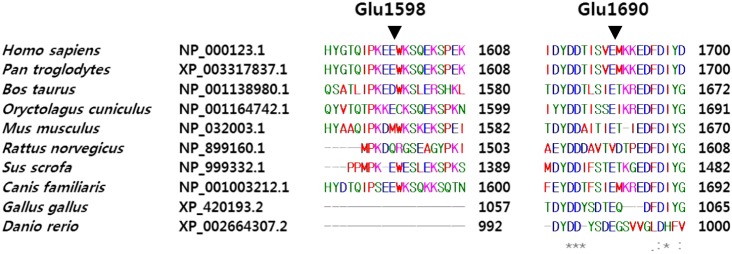
Thirteen individuals (13%) were hemizygous for sequence variations in the coding region of F8. Six (6%) had c.3780C>G (p.Asp1260Glu), five (5%) had c.3864A>C (p.Ser1288=). One each individual (1%) had c.4794G>T (p.Glu1598Asp) and c.5069 A>G (p.Glu1690Gly). Asp1260Glu and Ser1288= were known SNPs (rs1800291 and rs1800292, respectively). Glu1598Asp was assigned as a missense mutation in public databases (HGMD and HAMSTeRS), and Glu1690Gly was a novel variation. Based on the normal FVIII activities in control individuals carrying these variations (109% and 148%, respectively), they were considered to be rare SNPs. No variation was observed in F9 of control individuals.
CONCLUSION
A significant proportion of control individuals carried sequence variations in F8, but not in F9. These results can be used as a reference dataset for molecular diagnosis of hemophilia A and B, particularly in Korea.
Keyword
MeSH Terms
Figure
Reference
-
1. GeneReviews: Hemophilia B. Seattle, WA: University of Washington;2011. Accessed February 25, 2013. at http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=hemo-b.2. GeneReviews: Hemophilia A. Seattle, WA: University of Washington;2011. Accessed February 25, 2013. at http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=hemo-a.3. dbSNP Short genetic variations. The NCBI Single Nucleotide Polymorphism database. Bethesda, MD: NCBI;2012. Accessed February 25, 2013. at http://www.ncbi.nlm.nih.gov/SNP/.4. Viel KR, Machiah DK, Warren DM, et al. A sequence variation scan of the coagulation factor VIII (FVIII) structural gene and associations with plasma FVIII activity levels. Blood. 2007; 109:3713–3724. PMID: 17209060.
Article5. Singer H, Ahmed R, Ivaskevicius V, et al. Genetic variability of the factor VIII gene in the normal population. In : 34th Hemophilia Symposium; 2005. p. 348–350.6. Machiah D, Viel K, Almasy L, et al. A common SNP in the factor VIII (f-VIII) gene encodes a conservative aspartate to glutamate substitution (Asp1241Glu) in the B-domain that influences f-VIII activity levels. Blood. 2003; 102(Suppl):abst 181.7. Scanavini D, Legnani C, Lunghi B, Mingozzi F, Palareti G, Bernardi F. The factor VIII D1241E polymorphism is associated with decreased factor VIII activity and not with activated protein C resistance levels. Thromb Haemost. 2005; 93:453–456. PMID: 15735794.
Article8. HAMSTeRS. The Haemophilia A Mutation, Structure, Test and Resource Site. London, UK: MRC Clinical Sciences Centre;2012. Accessed February 25, 2013. at http://hadb.org.uk/.9. Grunebaum L, Cazenave JP, Camerino G, et al. Carrier detection of Hemophilia B by using a restriction site polymorphism associated with the coagulation Factor IX gene. J Clin Invest. 1984; 73:1491–1495. PMID: 6325506.
Article10. McCurdy PR. Use of genetic linkage for the detection of female carriers of hemophilia. N Engl J Med. 1971; 285:218–219. PMID: 5087725.
Article11. Oberle I, Camerino G, Heilig R, et al. Genetic screening for hemophilia A (classic hemophilia) with a polymorphic DNA probe. N Engl J Med. 1985; 312:682–686. PMID: 2983207.
Article12. Liu Q, Nozari G, Sommer SS. Single-tube polymerase chain reaction for rapid diagnosis of the inversion hotspot of mutation in hemophilia A. Blood. 1998; 92:1458–1459. PMID: 9694739.
Article13. Keeney S, Watson P, Hay C, Cumming A. Rapid turnaround for full mutation analysis of larger genes: robotic processing and automated DNA sequencing of the essential regions of the FVIII (F8) gene for mutation identification in haemophilia A. J Med Genet. 2004; 41(Suppl 1):S67.14. Rossetti LC, Radic CP, Larripa IB, De Brasi CD. Genotyping the hemophilia inversion hotspot by use of inverse PCR. Clin Chem. 2005; 51:1154–1158. PMID: 15860568.
Article15. Kwon MJ, Yoo KY, Kim HJ, Kim SH. Identification of mutations in the F9 gene including exon deletion by multiplex ligationdependent probe amplification in 33 unrelated Korean patients with haemophilia B. Haemophilia. 2008; 14:1069–1075. PMID: 18624698.16. Kim HJ, Chung HS, Kim SK, et al. Mutation spectrum and inhibitor risk in 100 Korean patients with severe haemophilia A. Haemophilia. 2012; 18:1008–1013. PMID: 22741565.
Article17. Seattle SNPs. National Heart, Lung, and Blood Institute Program for genomic applications. Bethesda, MD: NHLBI;2009. Accessed February 25, 2013. at http://pga.gs.washington.edu.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of a shared F8 mutation in the Korean patients with acquired hemophilia A
- Usefulness of HhaI and MseI DNA Polymorphism of Factor IX Gene in the Molecular Genetic Diagnosis of Hemophilia B in Korean Population
- Direct detection of hemophilia B F9 gene mutation using multiplex PCR and conformation sensitive gel electrophoresis
- Hemophilia
- Molecular genetic study of St14.1(DXS52) TaqI RFLPs in Koreans for the diagnosis of hemophilia A