Allergy Asthma Immunol Res.
2014 Jul;6(4):333-340. 10.4168/aair.2014.6.4.333.
Gene - Gene Interactions Among MCP Genes Polymorphisms in Asthma
- Affiliations
-
- 1Respiratory and Allergy Medicine, Interanl Medicine, Soonchunhyang University Bucheon Hospital, Bucheon, Korea. mdcspark@unitel.co.kr
- KMID: 2260214
- DOI: http://doi.org/10.4168/aair.2014.6.4.333
Abstract
- PURPOSE
Monocyte chemoattractant proteins (MCPs) are important cytokines that involved in cellular activation and releasing of inflammatoy mediators by basophils and eosinophils in allergic disease. Some MCP gene variants implicate in asthma and monoclonal antibody for MCP-3 blocks allergic inflammations in the patients with asthma. Detection of interactions between gene and environment or between genes for complex disease such as asthma is important. We searched for an evidence of genetic effect of single nucleotide polymorphisms (SNPs) of MCP genes as well as gene - gene interactions involved in asthma.
METHODS
Four hundreds asthmatics and four hundreds normal controls were enrolled. Asthma was defined as a positive bronchodilator response or positive methacholine provocation test with compatible clinical symptoms. Seven MCP gene SNPs (2 SNPs in MCP-1, 1 in MCP-2, and 4 in MCP-3) were included. Association analyses between SNP and asthma, and the tests for gene - gene interaction were performed.
RESULTS
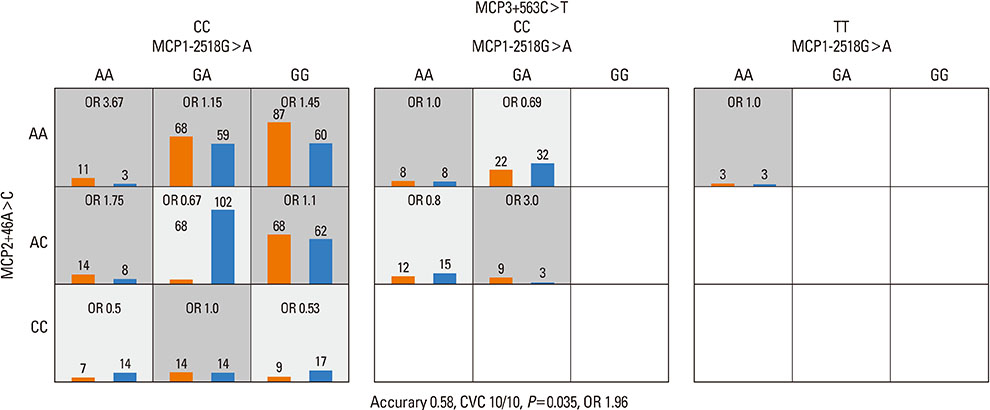
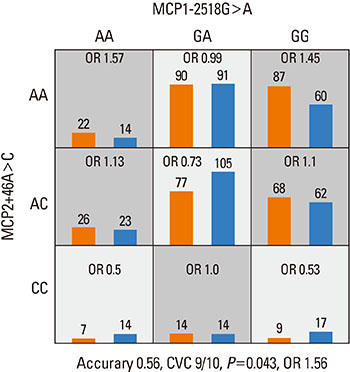
Strong linkage disequilibria were found among 7 MCP gene polymorphisms. There was no SNP that showed a significant association with asthma among 7 SNPs of 3 MCP genes. No haplotype was associated with asthma, either. The combination of MCP1-2518G>A, MCP2+46A>C, and MCP3+563C>T was the best predictive model for asthma as compared to the control in tests for gene - gene interaction. The MCP1-2518G>A and MCP2+46A>C was the second best predictive combination and this had the highest synergistic interaction effect on the subject's status than any other combination of polymorphisms. Complete linkages were not associated with the gene - gene interactions models.
CONCLUSIONS
MCP gene polymorphisms probably interact with each other; thus, these findings may help in developing a possible genetic marker to predict asthma.
MeSH Terms
Figure
Reference
-
1. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012; 18:716–725.2. Holgate ST. A brief history of asthma and its mechanisms to modern concepts of disease pathogenesis. Allergy Asthma Immunol Res. 2010; 2:165–171.3. Santiago J, Hernández-Cruz JL, Manjarrez-Zavala ME, Montes-Vizuet R, Rosete-Olvera DP, Tapia-Díaz AM, Zepeda-Peney H, Terán LM. Role of monocyte chemotactic protein-3 and -4 in children with virus exacerbation of asthma. Eur Respir J. 2008; 32:1243–1249.4. Sozzani S, Zhou D, Locati M, Rieppi M, Proost P, Magazin M, Vita N, van Damme J, Mantovani A. Receptors and transduction pathways for monocyte chemotactic protein-2 and monocyte chemotactic protein-3. Similarities and differences with MCP-1. J Immunol. 1994; 152:3615–3622.5. Pype JL, Dupont LJ, Menten P, Van Coillie E, Opdenakker G, Van Damme J, Chung KF, Demedts MG, Verleden GM. Expression of monocyte chemotactic protein (MCP)-1, MCP-2, and MCP-3 by human airway smooth-muscle cells. Modulation by corticosteroids and T-helper 2 cytokines. Am J Respir Cell Mol Biol. 1999; 21:528–536.6. Holgate ST, Bodey KS, Janezic A, Frew AJ, Kaplan AP, Teran LM. Release of RANTES, MIP-1 alpha, and MCP-1 into asthmatic airways following endobronchial allergen challenge. Am J Respir Crit Care Med. 1997; 156:1377–1383.7. Chan CK, Kuo ML, Yeh KW, Ou LS, Chen LC, Yao TC, Huang JL. Sequential evaluation of serum monocyte chemotactic protein 1 among asymptomatic state and acute exacerbation and remission of asthma in children. J Asthma. 2009; 46:225–228.8. Campbell EM, Charo IF, Kunkel SL, Strieter RM, Boring L, Gosling J, Lukacs NW. Monocyte chemoattractant protein-1 mediates cockroach allergen-induced bronchial hyperreactivity in normal but not CCR2-/- mice: the role of mast cells. J Immunol. 1999; 163:2160–2167.9. Powell N, Humbert M, Durham SR, Assoufi B, Kay AB, Corrigan CJ. Increased expression of mRNA encoding RANTES and MCP-3 in the bronchial mucosa in atopic asthma. Eur Respir J. 1996; 9:2454–2460.10. Humbert M, Ying S, Corrigan C, Menz G, Barkans J, Pfister R, Meng Q, Van Damme J, Opdenakker G, Durham SR, Kay AB. Bronchial mucosal expression of the genes encoding chemokines RANTES and MCP-3 in symptomatic atopic and nonatopic asthmatics: relationship to the eosinophil-active cytokines interleukin (IL)-5, granulocyte macrophage-colony-stimulating factor, and IL-3. Am J Respir Cell Mol Biol. 1997; 16:1–8.11. Szalai C, Kozma GT, Nagy A, Bojszkó A, Krikovszky D, Szabó T, Falus A. Polymorphism in the gene regulatory region of MCP-1 is associated with asthma susceptibility and severity. J Allergy Clin Immunol. 2001; 108:375–381.12. Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004; 429:446–452.13. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, November 1986. Am Rev Respir Dis. 1987; 136:225–244.14. Crapo RO, Casaburi R, Coates AL, Enright PL, Hankinson JL, Irvin CG, MacIntyre NR, McKay RT, Wanger JS, Anderson SD, Cockcroft DW, Fish JE, Sterk PJ. Guidelines for methacholine and exercise challenge testing-1999. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, July 1999. Am J Respir Crit Care Med. 2000; 161:309–329.15. Ferris BG. Epidemiology standardization project (American Thoracic Society). Am Rev Respir Dis. 1978; 118:1–120.16. Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987; 117:331–341.17. Ding K, Zhou K, He F, Shen Y. LDA--a java-based linkage disequilibrium analyzer. Bioinformatics. 2003; 19:2147–2148.18. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001; 68:978–989.19. Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001; 69:138–147.20. Lee JH, Moore JH, Park SW, Jang AS, Uh ST, Kim YH, Park CS, Park BL, Shin HD. Genetic interactions model among Eotaxin gene polymorphisms in asthma. J Hum Genet. 2008; 53:867–875.21. Lee JH, Jang AS, Park SW, Kim DJ, Park CS. Gene-gene interaction between CCR3 and eotaxin genes: the relationship with blood eosinophilia in asthma. Allergy Asthma Immunol Res. 2014; 6:55–60.22. Greene CS, Himmelstein DS, Nelson HH, Kelsey KT, Williams SM, Andrew AS, Karagas MR, Moore JH. Enabling personal genomics with an explicit test of epistasis. Pac Symp Biocomput. 2010; 327–336.23. Chung Y, Lee SY, Elston RC, Park T. Odds ratio based multifactor-dimensionality reduction method for detecting gene-gene interactions. Bioinformatics. 2007; 23:71–76.24. Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006; 27:1030–1037.25. Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006; 241:252–261.26. Lee JH, Choi JH, Yoon JA, Park SW, Kim D, Jang AS, Park JS, Uh ST, Rhim T, Kim YH, Shin HD, Park B, Park CS. An interaction model of IL-4 and IFN-γ gene SNPs in relation to atopy. Korean J Asthma Allergy Clin Immunol. 2008; 28:45–52.27. Jakulin A, Bratko I, Smrke D, Demšar J, Zupan B. Attribute interactions in medical data analysis. In : Dojat M, Keravnou-Papailiou E, Barahona P, editors. Artificial intelligence in medicine. Vol. 2780. Lecture notes in computer science. New York (NY): Springer;2003. p. 229–238.28. Curk T, Demsar J, Xu Q, Leban G, Petrovic U, Bratko I, Shaulsky G, Zupan B. Microarray data mining with visual programming. Bioinformatics. 2005; 21:396–398.29. Keszei M, Nagy A, Kozma GT, Radosits K, Tölgyesi G, Falus A, Szalai C. Pediatric asthmatic patients have low serum levels of monocyte chemoattractant protein-1. J Asthma. 2006; 43:399–404.30. Park BL, Kim LH, Choi YH, Cheong HS, Park HS, Hong SJ, Choi BW, Lee JH, Uh ST, Park CS, Shin HD. Association analysis of monocyte chemotactic protein-3 (MCP3) polymorphisms with asthmatic phenotypes. J Biochem Mol Biol. 2005; 38:77–81.31. Brodie ED. Why evolutionary genetics does not always add up. In : Wolf JB, Brodie ED, Wade MJ, editors. Epistasis and the evolutionary process. New York (NY): Oxford University Press;2000. p. 3–19.32. Yao TC, Wu KC, Chung HT, Shaw CK, Kuo ML, Wu CJ, Huang JL. MCP-1 gene regulatory region polymorphism in Chinese children with mild, moderate and near-fatal asthma. Allergy. 2004; 59:436–441.33. Belperio JA, Keane MP, Burdick MD, Lynch JP 3rd, Xue YY, Berlin A, Ross DJ, Kunkel SL, Charo IF, Strieter RM. Critical role for the chemokine MCP-1/CCR2 in the pathogenesis of bronchiolitis obliterans syndrome. J Clin Invest. 2001; 108:547–556.34. Navratilova Z, Mrazek F, Kriegova E, Hutyrova B, Kolek V, du Bois RM, Petrek M. The MCP-1-2518 (A to G) single nucleotide polymorphism in Czech patients with pulmonary sarcoidosis: association with Löfgren's syndrome. Sarcoidosis Vasc Diffuse Lung Dis. 2007; 24:33–38.35. Alonso-Villaverde C, Coll B, Parra S, Montero M, Calvo N, Tous M, Joven J, Masana L. Atherosclerosis in patients infected with HIV is influenced by a mutant monocyte chemoattractant protein-1 allele. Circulation. 2004; 110:2204–2209.36. Kaur S, Panicker SR, James T, Sarma PS, Thankappan KR, Kartha CC. Association of monocyte chemoattractant protein-1 -2518 polymorphism with metabolic syndrome in a South Indian cohort. Metab Syndr Relat Disord. 2009; 7:193–198.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gene-Gene Interaction Between CCR3 and Eotaxin Genes: The Relationship With Blood Eosinophilia in Asthma
- Severe Refractory Asthma and Genetics
- Gene-Environment Interactions Should be Considered in Future Studies to Understand the Association Between Prenatal Folate Supplementation and Asthma Development
- Association study of polymorphism in leukotriene C4 synthase and cysteinyl leukotriene receptor 1 genes with phenotype of asthma and clinical parameters in Korean children
- Gene-Environment Interactions in Asthma: Genetic and Epigenetic Effects