J Bacteriol Virol.
2014 Jun;44(2):162-169. 10.4167/jbv.2014.44.2.162.
Microbial Profile of the Stomach: Comparison between Normal Mucosa and Cancer Tissue in the Same Patient
- Affiliations
-
- 1Department of Microbiology, School of Medicine, Keimyung University, Daegu, Korea. wonki@dsmc.or.kr
- KMID: 2168691
- DOI: http://doi.org/10.4167/jbv.2014.44.2.162
Abstract
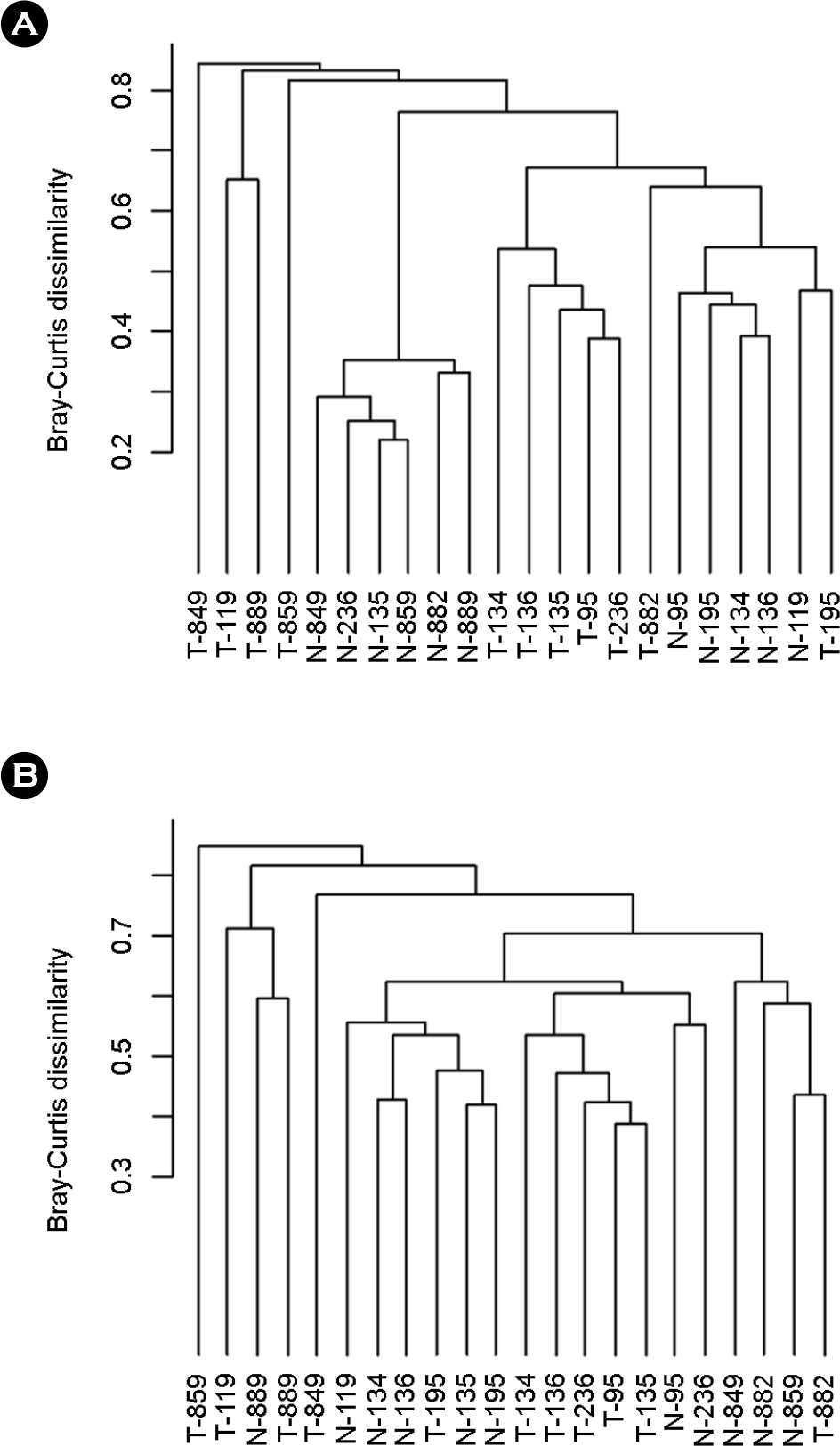
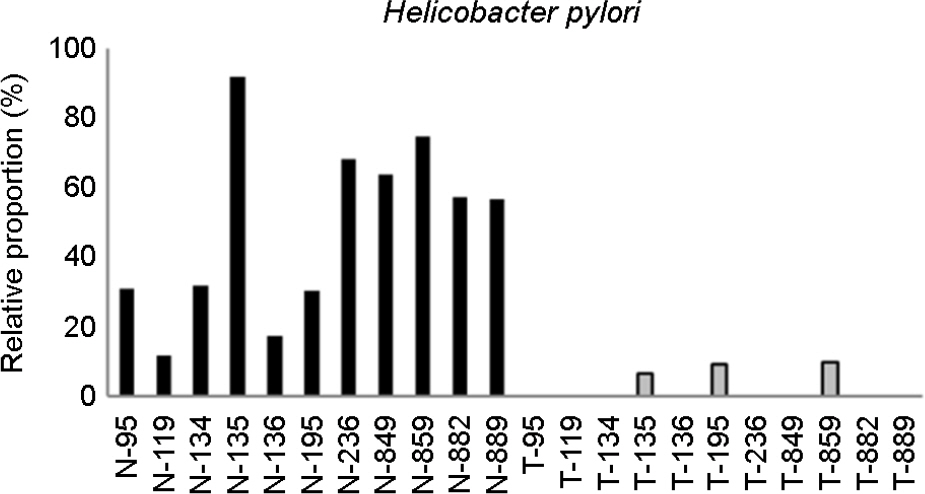
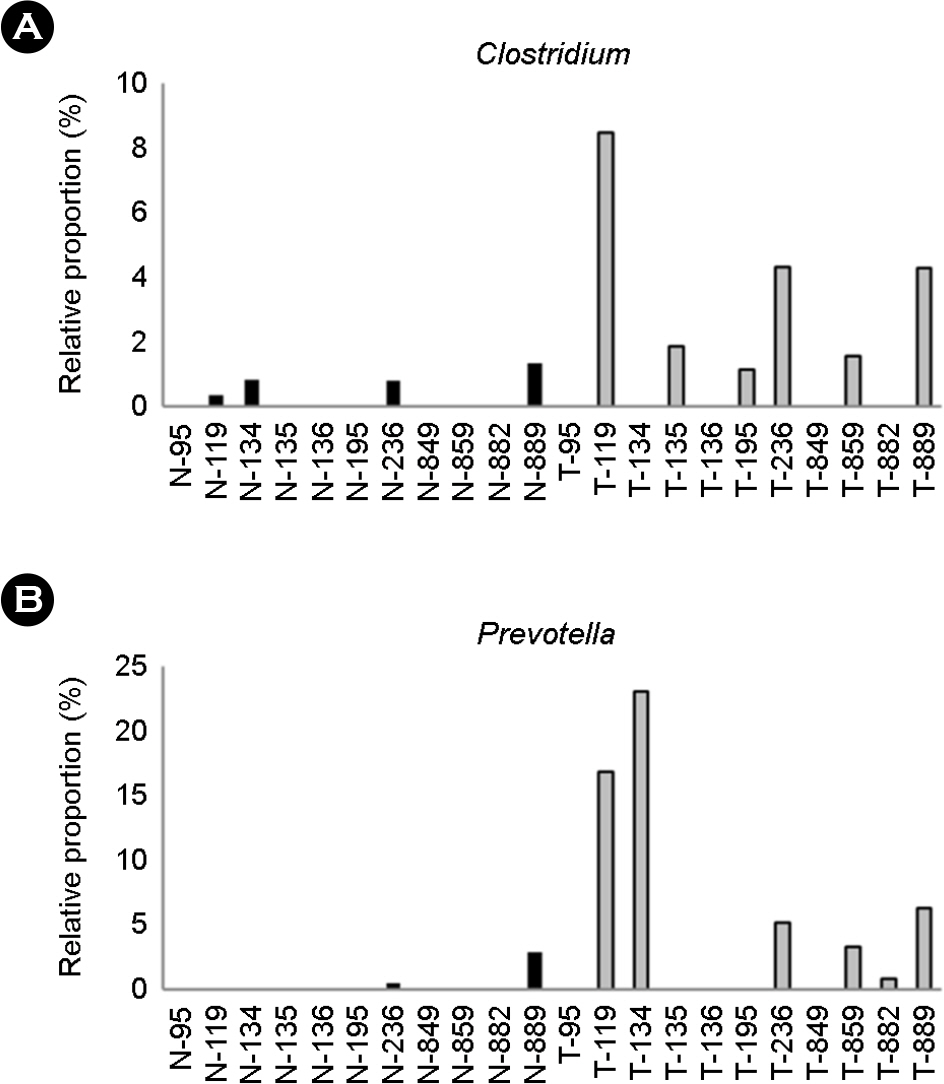
- Gastric cancer is the third most common cancer and the third most frequent cause of cancer mortality in Asia. It is predicted that gastric cancer will remain an important cause of death at least during the next half century because of the increasing number of new cases in an aging population. However, little has been revealed about the role of gastric microbes and their reaction to gastric cancer. In this study, we identified differences in the microbial communities between gastric cancer and normal gastric mucosa by comparing the microbiomes of tissues from the same patients. The clustering analysis results showed different bacterial communities between normal gastric mucosa and gastric cancer. A comparison of bacterial communities at the species level revealed that Helicobacter pylori was significantly reduced in cancer tissue compared to that in normal gastric mucosa in the same patient. A comparison at the genus level showed that Propionibacterium spp., Staphylococcus spp., and Corynebacterium spp. had significantly reduced populations in cancer tissue, whereas Clostridium spp. and Prevotella spp. had significantly increased populations in cancer tissue.
Keyword
MeSH Terms
Figure
Reference
-
1). Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013; 37:736–61.
Article2). No authors listed. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7∼14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994; 61:1–241.3). Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991; 325:1127–31.4). Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006; 103:732–7.
Article5). Hu Y, He LH, Xiao D, Liu GD, Gu YX, Tao XX, et al. Bacterial flora concurrent with Helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J Gastroenterol. 2012; 18:1257–61.6). Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011; 5:574–9.7). Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011; 1:170.
Article8). Albarracin VH, Pathak GP, Douki T, Cadet J, Borsarelli CD, Gärtner W, et al. Extremophilic Acinetobacter strains from high-altitude lakes in Argentinean Puna: remarkable UV-B resistance and efficient DNA damage repair. Orig Life Evol Biosph. 2012; 42:201–21.9). Wroblewski LE, Peek RM Jr. Helicobacter pylori in gastric carcinogenesis: mechanisms. Gastroenterol Clin North Am. 2013; 42:285–98.10). Wang ZK, Yang YS. Upper gastrointestinal microbiota and digestive diseases. World J Gastroenterol. 2013; 19:1541–50.
Article11). Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol. 2013.
Article12). Williams C, McColl KE. Review article: proton pump inhibitors and bacterial overgrowth. Aliment Pharmacol Ther. 2006; 23:3–10.
Article13). Osaki T, Matsuki T, Asahara T, Zaman C, Hanawa T, Yonezawa H, et al. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb Pathog. 2012; 53:12–8.14). Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009; 58:509–16.
Article15). Yoon K, Lee S, Han TS, Moon SY, Yun SM, Kong SH, et al. Comprehensive genome- and transcriptome-wide analyses of mutations associated with microsatellite instability in Korean gastric cancers. Genome Res. 2013; 23:1109–17.
Article16). Isakov O, Modai S, Shomron N. Pathogen detection using short-RNA deep sequencing subtraction and assembly. Bioinformatics. 2011; 27:2027–30.
Article17). Xu Y, Stange-Thomann N, Weber G, Bo R, Dodge S, David RG, et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics. 2003; 81:329–35.
Article18). Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–11.
Article19). Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008; 18:821–9.
Article20). Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. vegan: Community Ecology Package. 2013. [cited 2014 Mar 3]. Available from:. http://CRAN.R-project.org/package=vegan/.21). Leja M, Wex T, Malfertheiner P. Markers for gastric cancer premalignant lesions: where do we go? Dig Dis. 2012; 30:268–76.
Article22). Yeh JM, Hur C, Schrag D, Kuntz KM, Ezzati M, Stout N, et al. Contribution of H. pylori and smoking trends to US incidence of intestinal-type noncardia gastric adenocarcinoma: a microsimulation model. PLoS Med. 2013; 10:e1001451.23). Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012; 30:434–9.
Article24). Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, et al. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007; 81:11332–40.
Article25). Rathi B, Sarangi AN, Trivedi N. Genome subtraction for novel target definition in Salmonella typhi. Bioinformation. 2009; 4:143–50.26). Lauwers GY, Srivastava A. Gastric preneoplastic lesions and epithelial dysplasia. Gastroenterol Clin North Am. 2007; 36:813–29.
Article27). Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008; 134:306–23.28). Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013; 11:385–99.29). Delgado S, Suárez A, Mayo B. Identification, typing and characterisation of Propionibacterium strains from healthy mucosa of the human stomach. Int J Food Microbiol. 2011; 149:65–72.30). Giannella RA, Broitman SA, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut. 1972; 13:251–6.31). Oh JD, Kling-Bäckhed H, Giannakis M, Engstrand LG, Gordon JI. Interactions between gastric epithelial stem cells and Helicobacter pylori in the setting of chronic atrophic gastritis. Curr Opin Microbiol. 2006; 9:21–7.32). Wu WM, Yang YS, Peng LH. Microbiota in stomach: New insights. J Dig Dis. 2014; 15:54–61.33). Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010; 10:785–94.
Article34). Tan MP, Kaparakis M, Galic M, Pedersen J, Pearse M, Wijburg OL, et al. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl Environ Microbiol. 2007; 73:1010–3.35). Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA, et al. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One. 2013; 8:e76375.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gastric Cancer and Non-Helicobacter pylori Microbiota
- A Study of beta-glucuronidase and Lactic Dehydrogenace Activities in the Neoplastic Tissue of Stomach Carcinoma
- Measurement of Telomerase Activity and Telomerase Reverse Transcriptase Expression in Gastric Fluid and Tissue for Early Diagnosis of Stomach Cancer
- Higher Microbial Abundance and Diversity in Bronchus-Associated Lymphoid Tissue Lymphomas Than in Non-cancerous Lung Tissues
- Comparison of c-erbB-2 Oncoprotein Expression in Serum and Tissue of Gastric Cancer Patients