J Bacteriol Virol.
2015 Jun;45(2):67-78. 10.4167/jbv.2015.45.2.67.
Glycopeptide Antibiotics: Structure and Mechanisms of Action
- Affiliations
-
- 1Department of Biomedical Sciences, Chosun University, Gwangju, Korea. y_k_park@chosun.ac.kr
- 2Research Center for Proteinaceous Materials, Chosun University, Gwangju, Korea.
- KMID: 2149207
- DOI: http://doi.org/10.4167/jbv.2015.45.2.67
Abstract
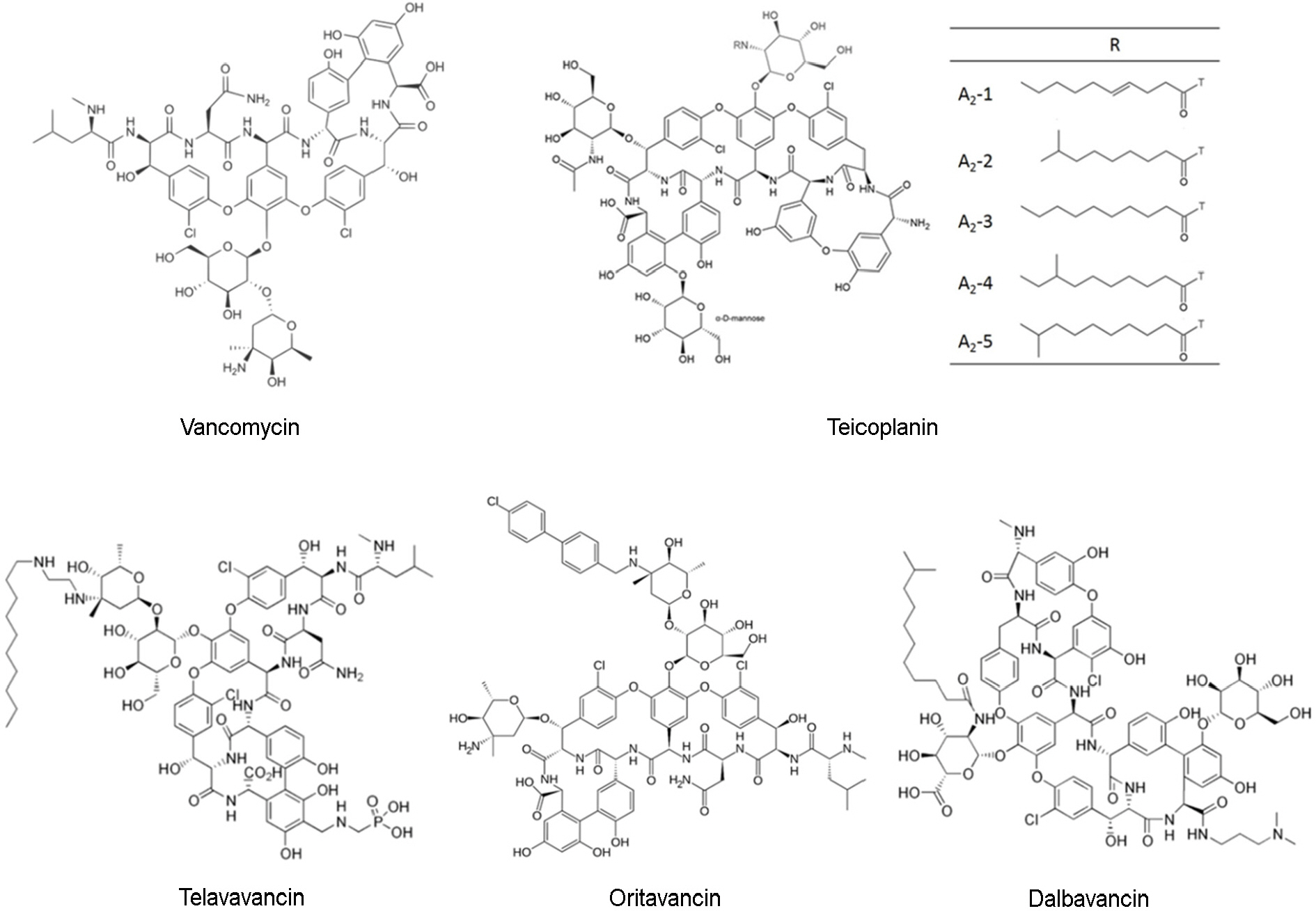
- Glycopeptides of the clinically important antibiotic drugs are glycosylated cyclic or polycyclic nonribosomal peptides. Glycopeptides such as vancomycin and teicoplanin are often used for the treatment of gram-positive bacteria in patients. The increased incidence of drug resistance and inadequacy of these therapeutics against gram-positive bacterial infections would be the formation and clinical development of more variable second generation of glycopeptide antibiotics: semisynthetic lipoglycopeptide analogs such as telavancin, dalbavancin, and oritavancin with improved activity and better pharmacokinetic properties. In this review, we describe the development of and bacterial resistance to vancomycin, teicoplanin, and semisynthetic glycopeptides (teicoplanin, dalbavancin, and oritavancin). The clinical influence of resistance to glycopeptides, particularly vancomycin, are also discussed.
Keyword
MeSH Terms
Figure
Reference
-
1). James RC, Pierce JG, Okano A, Xie J, Boger DL. Redesign of glycopeptide antibiotics: back to the future. ACS Chem Biol. 2012; 7:797–804.
Article2). Butler MS, Hansford KA, Blaskovich MA, Halai R, Cooper MA. Glycopeptide antibiotics: back to the future. J Antibiot (Tokyo). 2014; 67:631–44.
Article3). Reynolds PE. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989; 8:943–50.
Article4). Binda E, Marinelli F, Marcone GL. Old and New glycopeptide antibiotics: action and resistance. Antibiotics. 2014; 3:572–94.
Article5). Marcone GL, Binda E, Carrano L, Bibb M, Marinelli F. Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob Agents Chemother. 2014; 58:5191–201.6). Rossolini GM, Arena F, Pollini S. Novel infectious diseases and emerging gram-positive multiresistant pathogens in hospital and community acquired infections. Antimicrobials; Marinelli F, Genilloud O, editors. 1st ed.Berlin Heidelberg: Springer Verlag;2014.7). Treviño J, Bayón C, Ardá A, Marinelli F, Gandolfi R, Molinari F, et al. New insights into glycopeptide antibiotic binding to cell wall precursors using SPR and NMR spectroscopy. Chemistry. 2014; 20:7363–72.
Article8). Allen NE, Nicas TI. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol Rev. 2003; 26:511–32.
Article9). Van Bambeke F, Van Laethem Y, Courvalin P, Tulkens PM. Glycopeptide antibiotics: from conventional molecules to new derivatives. Drugs. 2004; 64:913–36.10). Parenti F, Beretta G, Berti M, Arioli V. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. I. Description of the producer strain, fermentation studies and biological properties. J Antibiot (Tokyo). 1978; 31:276–83.11). Sharma V, Jindal N. In vitro activity of vancomycin and teicoplanin against coagulase negative staphylococci. Oman Med J. 2011; 26:186–8.12). Marcone GL, Marinelli F. Glycopeptides: An old but up to date successful antibiotic class. Antimicrobials; Marinelli F, Genilloud O, editors. 1st ed.Berlin Heidelberg: Springer Verlag;2014.13). Cooper MA, Williams DH. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem Biol. 1999; 6:891–9.
Article14). Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005; 105:425–48.
Article15). Zhanel GG, Calic D, Schweizer F, Zelenitsky S, Adam H, Lagacé-Wiens PR, et al. New lipoglycopeptides: A comparative review of dalbavancin, oritavancin and telavancin. Drugs. 2010; 70:859–86.16). Ashford PA, Bew SP. Recent advances in the synthesis of new glycopeptide antibiotics. Chem Soc Rev. 2012; 41:957–78.
Article17). Leadbetter MR, Adams SM, Bazzini B, Fatheree PR, Karr DE, Krause KM, et al. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424). J Antibiot (Tokyo). 2004; 57:326–36.
Article18). Shaw JP, Seroogy J, Kaniga K, Higgins DL, Kitt M, Barriere S. Pharmacokinetics, serum inhibitory and bactericidal activity, and safety of telavancin in healthy subjects. Antimicrob Agents Chemother. 2005; 49:195–201.
Article19). Sun HK, Duchin K, Nightingale CH, Shaw JP, Seroogy J, Nicolau DP. Tissue penetration of telavancin after intravenous administration in healthy subjects. Antimicrob Agents Chemother. 2006; 50:788–90.
Article20). Zhanel GG, Trapp S, Gin AS, DeCorby M, Lagacé-Wiens PR, Rubinstein E, et al. Dalbavancin and telavancin: novel lipoglycopeptides for the treatment of Gram-positive infections. Expert Rev Anti Infect Ther. 2008; 6:67–81.
Article21). Das B, Sarkar C, Schachter J. Oritavancin. – a new semisynthetic lipoglycopeptide agent to tackle the challenge of resistant gram positive pathogens. Pak J Pharm Sci. 2013; 26:1045–55.22). Zhanel GG, Schweizer F, Karlowsky JA. Oritavancin: mechanism of action. Clin Infect Dis. 2012; 54:S214–9.
Article23). Belley A, McKay GA, Arhin FF, Sarmiento I, Beaulieu S, Fadhil I, et al. Oritavancin disrupts membrane integrity of Staphylococcus aureus and vancomycin-resistant enterococci to effect rapid bacterial killing. Antimicrob Agents Chemother. 2010; 54:5369–71.24). Malabarba A, Goldstein BP. Origin, structure, and activity in vitro and in vivo of dalbavancin. J Antimicrob Chemother. 2005; 55:ii15–20.25). Banik JJ, Brady SF. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr Opin Microbiol. 2010; 13:603–9.
Article26). Loll PJ, Axelsen PH. The structural biology of molecular recognition by vancomycin. Annu Rev Biophys Biomol Struct. 2000; 29:265–89.
Article27). Yang Z, Vorpagel ER, Laskin J. Influence of the charge state on the structures and interactions of vancomycin antibiotics with cell-wall analogue peptides: experimental and theoretical studies. Chemistry. 2009; 15:2081–90.
Article28). Belley A, Neesham-Grenon E, McKay G, Arhin FF, Harris R, Beveridge T, et al. Oritavancin kills stationary-phase and biofilm Staphylococcus aureus cells in vitro. Antimicrob Agents Chemother. 2009; 53:918–25.29). Saravolatz LD, Stein GE, Johnson LB. Telavancin: a novel lipoglycopeptide. Clin Infect Dis. 2009; 49:1908–14.
Article30). Bennett JW, Lewis JS, Ellis MW. Dalbavancin in the treatment of complicated skin and soft-tissue infections: a review. Ther Clin Risk Manag. 2008; 4:31–40.31). Sujatha S, Praharaj I. Glycopeptide resistance in gram-positive cocci: a review. Interdiscip Perspect Infect Dis. 2012; 2012:781679.
Article32). Arthur M, Quintiliani R Jr. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 2001; 45:375–81.
Article33). Wright GD, Holman TR, Walsh CT. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM-4147. Biochemistry. 1993; 32:5057–63.34). Healy VL, Lessard IA, Roper DI, Knox JR, Walsh CT. Vancomycin resistance in enterococci: reprogramming of the D-Ala-D-Ala ligases in bacterial peptidoglycan biosynthesis. Chem Biol. 2000; 7:R109–19.
Article35). Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006; 42:S25–34.
Article36). Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996; 4:401–7.
Article37). Depardieu F, Kolbert M, Pruul H, Bell J, Courvalin P. VanD-type vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2004; 48:3892–904.38). Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother. 2010; 54:4643–7.39). Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, et al. D-Ala-D-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011; 55:4606–12.40). Nomura T, Tanimoto K, Shibayama K, Arakawa Y, Fujimoto S, Ike Y, et al. Identification of VanN-type vancomycin resistance in an Enterococcus faecium isolate from chicken meat in Japan. Antimicrob Agents Chemother. 2012; 56:6389–92.41). Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991; 30:10408–15.42). Bugg TD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Identification of vancomycin resistance protein VanA as a D-alanine: D-alanine ligase of altered substrate specificity. Biochemistry. 1991; 30:2017–21.43). Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007; 20:79–114.
Article44). Yim G, Thaker MN, Koteva K, Wright G. Glycopeptide antibiotic biosynthesis. J Antibiot (Tokyo). 2014; 67:31–41.
Article45). Arthur M, Depardieu F, Snaith HA, Reynolds PE, Courvalin P. Contribution of VanY D, D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994; 38:1899–903.46). Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995; 154:87–92.47). Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992; 174:2582–91.48). Podmore AH, Reynolds PE. Purification and characterization of VanXY(C), a D, D-dipeptidase/D, D-carboxypeptidase in vancomycin-resistant Enterococcus gallinarum BM4174. Eur J Biochem. 2002; 269:2740–6.49). Smith TL, Pearson ML, Wilcox KR, Cruz C, Lancaster MV, Robinson-Dunn B, et al. Emergence of vancomycin resistance in Staphylococcus aureus. glycopeptide-intermediate Staphylococcus aureus working group. N Engl J Med. 1999; 340:493–501.50). Biavasco F, Manso E, Varaldo PE. In vitro activities of ramoplanin and four glycopeptide antibiotics against clinical isolates of Clostridium difficile. Antimicrob Agents Chemother. 1991; 35:195–7.51). Renzoni A, Kelley WL, Vaudaux P, Cheung AL, Lew DP. Exploring innate glycopeptide resistance mechanisms in Staphylococcus aureus. Trends Microbiol. 2010; 18:55–6.52). Finan JE, Archer GL, Pucci MJ, Climo MW. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001; 45:3070–5.53). Muthaiyan A, Jayaswal RK, Wilkinson BJ. Intact mutS in laboratory-derived and clinical glycopeptide-intermediate Staphylococcus aureus strains. Antimicrob Agents Chemother. 2004; 48:623–5.54). Appelbaum PC. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2006; 12:16–23.55). Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010; 23:99–139.56). Sakoulas G, Eliopoulos GM, Fowler VG Jr, Moellering RC Jr, Novick RP, Lucindo N, et al. Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob Agents Chemother. 2005; 49:2687–92.57). Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering Jr RC, Eliopoulos GM. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis. 2009; 199:532–6.58). Périchon B, Courvalin P. Heterologous expression of the enterococcal vanA operon in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2004; 48:4281–5.59). Van Bambeke F, Mingeot-Leclercq MP, Struelens MJ, Tulkens PM. The bacterial envelope as a target for novel anti-MRSA antibiotics. Trends Pharmacol Sci. 2008; 29:124–34.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Appropriate Use of Glycopeptide Antibiotics and Therapeutic Drug Monitoring for Invasive Infections
- Antimicrobial resistance in enterococci
- Drug Discovery Perspectives of Antisense Oligonucleotides
- A Case of Septicemia by Transient Glycopeptide: Dependent Enterococcus faecium
- Clinical Characteristics and Outcomes of Patients with Culture-Negative Pyogenic Spondylitis according to Empiric Glycopeptide Use