Tuberc Respir Dis.
2012 Jul;73(1):11-21. 10.4046/trd.2012.73.1.11.
Promoter Methylation of CDKN2A, RARbeta, and RASSF1A in Non-Small Cell Lung Carcinoma: Quantitative Evaluation Using Pyrosequencing
- Affiliations
-
- 1Department of Pathology, St. Mary's Hospital, The Catholic University of Korea School of Medicine, Daejeon, Korea. sulpark@freechal.com
- 2Department of Internal Medicine, Konyang University Hospital, Konyang University College of Medicine, Daejeon, Korea. sk1609@hanmail.net
- KMID: 2050670
- DOI: http://doi.org/10.4046/trd.2012.73.1.11
Abstract
- BACKGROUND
While qualitative analysis of methylation has been reviewed, the quantitative analysis of methylation has rarely been studied. We evaluated the methylation status of CDKN2A, RARbeta, and RASSF1A promoter regions in non-small cell lung carcinomas (NSCLCs) by using pyrosequencing. Then, we evaluated the association between methylation at the promoter regions of these tumor suppressor genes and the clinicopathological parameters of the NSCLCs.
METHODS
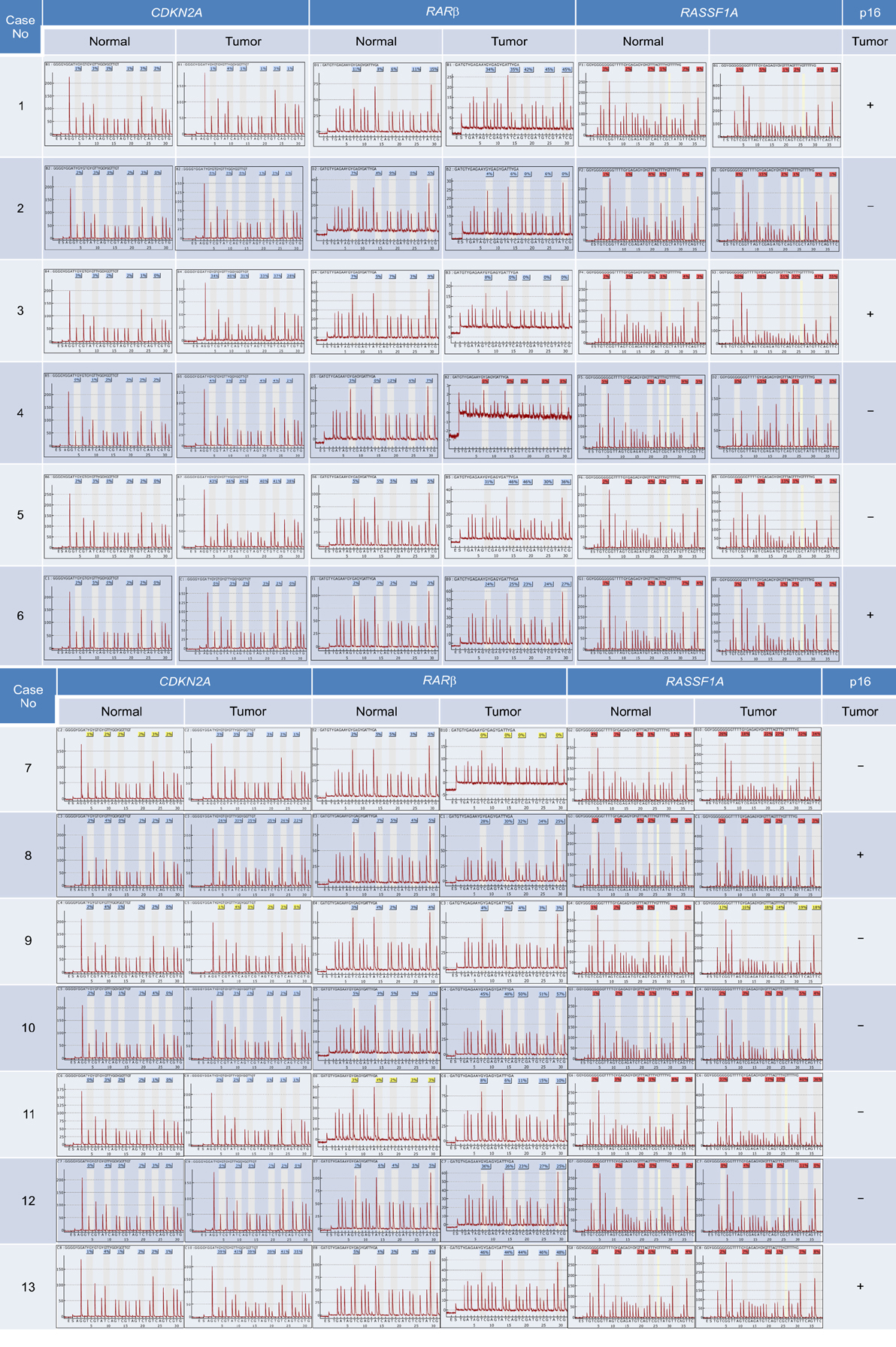
We collected tumor tissues from a total of 53 patients with NSCLCs and analyzed the methylation level of the CDKN2A, RARbeta, and RASSF1A promoter regions by using pyrosequencing. In addition, we investigated the correlation between the hypermethylation of CDKN2A and the loss of p16INK4A immunoexpression.
RESULTS
Hypermethylation of CDKN2A, RARbeta, and RASSF1A promoter regions were 16 (30.2%), 22 (41.5%), and 21 tumors (39.6%), respectively. The incidence of hypermethylation at the CDKN2A promoter in the tumors was higher in undifferentiated large cell carcinomas than in other subtypes (p=0.002). Hyperrmethylation of CDKN2A was significantly associated with p16INK4A immunoexpression loss (p=0.045). With regard to the clinicopathological characteristics of NSCLC, certain histopathological subtypes were found to be strongly associated with the loss of p16INK4A immunoexpression (p=0.016). Squamous cell carcinoma and undifferentiated large cell carcinoma showed p16INK4A immunoexpression loss more frequently. The Kaplan-Meier survival curves analysis showed that methylation level and patient survival were barely related to one another.
CONCLUSION
We quantitatively analyzed the promoter methylation status by using pyrosequencing. We showed a significant correlation between CDKN2A hypermethylation and p16INK4A immunoexpression loss.
Keyword
MeSH Terms
-
Carcinoma, Large Cell
Carcinoma, Non-Small-Cell Lung
Carcinoma, Squamous Cell
DNA Methylation
Evaluation Studies as Topic
Genes, p16
Genes, Tumor Suppressor
Humans
Incidence
Kaplan-Meier Estimate
Lung
Methylation
Promoter Regions, Genetic
Receptors, Retinoic Acid
Sequence Analysis, DNA
Tumor Suppressor Proteins
Receptors, Retinoic Acid
Tumor Suppressor Proteins
Figure
Reference
-
1. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998. 72:141–196.2. Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, et al. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995. 1:686–692.3. Buckingham L, Penfield Faber L, Kim A, Liptay M, Barger C, Basu S, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010. 126:1630–1639.4. Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999. 8:459–470.5. Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010. 11:137.6. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008. 359:1367–1380.7. Visbal AL, Williams BA, Nichols FC 3rd, Marks RS, Jett JR, Aubry MC, et al. Gender differences in non-small-cell lung cancer survival: an analysis of 4,618 patients diagnosed between 1997 and 2002. Ann Thorac Surg. 2004. 78:209–215.8. Koh J, Enders GH, Dynlacht BD, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995. 375:506–510.9. Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001. 61:3419–3424.10. Danesi R, de Braud F, Fogli S, de Pas TM, Di Paolo A, Curigliano G, et al. Pharmacogenetics of anticancer drug sensitivity in non-small cell lung cancer. Pharmacol Rev. 2003. 55:57–103.11. Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, Motoyama T. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003. 94:589–592.12. Brabender J, Usadel H, Metzger R, Schneider PM, Park J, Salonga D, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res. 2003. 9:223–227.13. Field JK, Liloglou T, Warrak S, Burger M, Becker E, Berlin K, et al. Methylation discriminators in NSCLC identified by a microarray based approach. Int J Oncol. 2005. 27:105–111.14. Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM, et al. Methylation of the 5 CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995. 55:4531–4535.15. Schmutte C, Jones PA. Involvement of DNA methylation in human carcinogenesis. Biol Chem. 1998. 379:377–388.16. El-Naggar AK, Lai S, Clayman G, Lee JK, Luna MA, Goepfert H, et al. Methylation, a major mechanism of p16/CDKN2 gene inactivation in head and neck squamous carcinoma. Am J Pathol. 1997. 151:1767–1774.17. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996. 93:9821–9826.18. Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol Biol. 2007. 373:89–102.19. González-Quevedo R, Iniesta P, Morán A, de Juan C, Sánchez-Pernaute A, Fernández C, et al. Cooperative role of telomerase activity and p16 expression in the prognosis of non-small-cell lung cancer. J Clin Oncol. 2002. 20:254–262.20. Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, et al. Promoter methylation of p16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer. 2006. 94:561–568.21. Shaw RJ, Hall GL, Lowe D, Liloglou T, Field JK, Sloan P, et al. The role of pyrosequencing in head and neck cancer epigenetics: correlation of quantitative methylation data with gene expression. Arch Otolaryngol Head Neck Surg. 2008. 134:251–256.22. Kontic M, Stojsic J, Jovanovic D, Bunjevacki V, Ognjanovic S, Kuriger J, et al. Aberrant promoter methylation of CDH13 and MGMT genes is associated with clinicopathologic characteristics of primary non-small-cell lung carcinoma. Clin Lung Cancer. 2012. 13:297–303.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Association between RASSF1A Methylation and Clinicopathological Factors in Patients with Squamous Cell Carcinoma of Lung
- Prognostic Role of Methylation Status of the MGMT Promoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients
- Clinical Significance of Aberrant Wnt7a Promoter Methylation in Human Non-Small Cell Lung Cancer in Koreans
- Quantitative analysis of COX-2 promoter methylation in gastric carcinoma
- Analysis of Epigenetic Marker of Bladder Cancer