J Korean Neurosurg Soc.
2016 Jan;59(1):26-36. 10.3340/jkns.2016.59.1.26.
Prognostic Role of Methylation Status of the MGMT Promoter Determined Quantitatively by Pyrosequencing in Glioblastoma Patients
- Affiliations
-
- 1Department of Pathology, Dong-A University College of Medicine, Busan, Korea.
- 2Department of Neurosurgery, Dong-A University College of Medicine, Busan, Korea.
- 3Division of Neurooncology, Department of Neurosurgery, Samsung Changwon Hospital, Sungkyunkwan University School of Medicine, Changwon, Korea. yzkim@skku.edu
- KMID: 2152915
- DOI: http://doi.org/10.3340/jkns.2016.59.1.26
Abstract
OBJECTIVE
This study investigated whether pyrosequencing can be used to determine the methylation status of the MGMT promoter as a clinical biomarker using relatively old archival tissue samples of glioblastoma. We also examined other prognostic factors for survival of glioblastoma patients.
METHODS
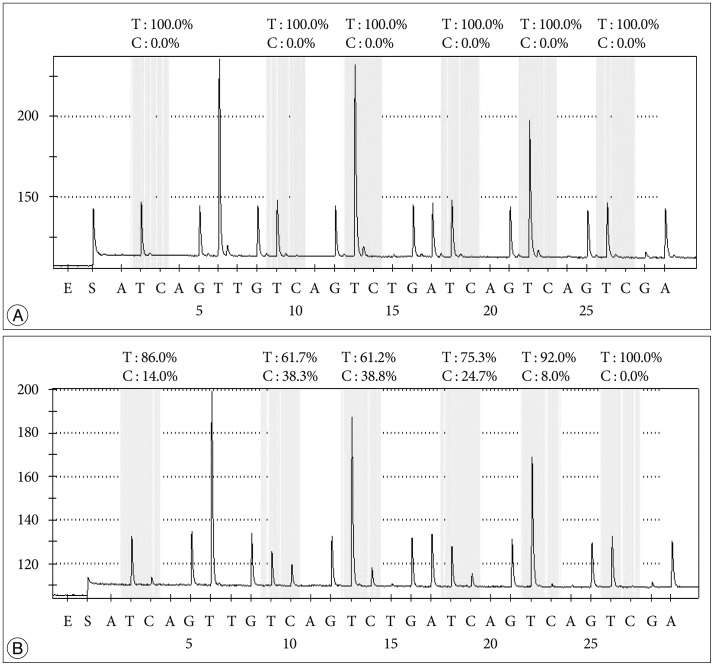
The available study set included formalin-fixed paraffin-embedded (FFPE) tissue from 104 patients at two institutes from 1997 to 2012, all of which were diagnosed histopathologically as glioblastoma. Clinicopathologic data were collected by review of medical records. For pyrosequencing analysis, the PyroMark Q96 CpG MGMT kit (Qiagen, Hilden, Germany) was used to detect the level of methylation at exon 1 positions 17-39 of the MGMT gene, which contains 5 CpGs.
RESULTS
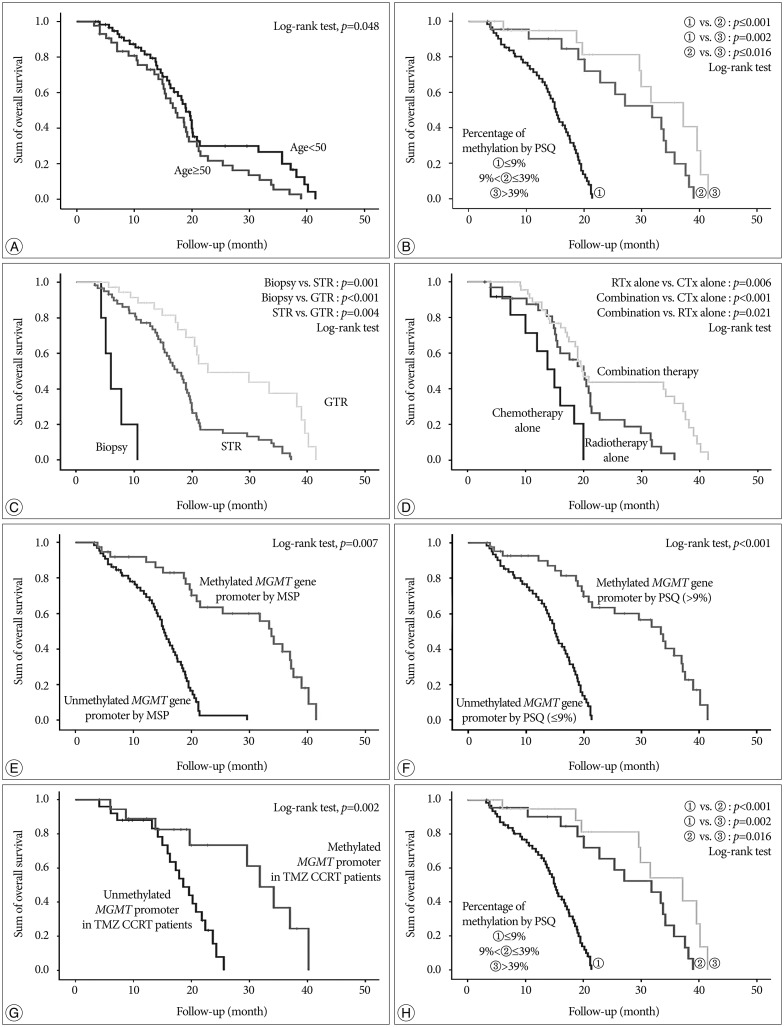
Methylation of the MGMT promoter was detected in 43 (41.3%) of 104 samples. The average percentage methylation was 14.0+/-16.8% overall and 39.0+/-14.7% for methylated cases. There was no significant pattern of linear increase or decrease according to the age of the FFPE block (p=0.687). In multivariate analysis, age, performance status, extent of surgery, method of adjuvant therapy, and methylation status estimated by pyrosequencing were independently associated with overall survival. Additionally, patients with a high level of methylation survived longer than those with low methylation (p=0.016).
CONCLUSION
In this study, the status and extent of methylation of the MGMT promoter analyzed by pyrosequencing were associated with overall survival in glioblastoma patients. Pyrosequencing is a quantitative method that overcomes the problems of MSP and a simple technique for accurate analysis of DNA sequences.
Keyword
MeSH Terms
Figure
Reference
-
1. Derks S, Lentjes MH, Hellebrekers DM, de Bruïne AP, Herman JG, van Engeland M. Methylation-specific PCR unraveled. Cell Oncol. 2004; 26:291–299. PMID: 15623939.
Article2. Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009; 101:124–131. PMID: 19536096.
Article3. Eng J. Receiver operating characteristic analysis : a primer. Acad Radiol. 2005; 12:909–916. PMID: 16039544.4. Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000; 343:1350–1354. PMID: 11070098.
Article5. Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, et al. DNA methylation in glioblastoma : impact on gene expression and clinical outcome. BMC Genomics. 2010; 11:701. PMID: 21156036.6. Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992; 89:1827–1831. PMID: 1542678.
Article7. Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005; 352:997–1003. PMID: 15758010.
Article8. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR : a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996; 93:9821–9826. PMID: 8790415.
Article9. Jung KW, Ha J, Lee SH, Won YJ, Yoo H. An updated nationwide epidemiology of primary brain tumors in republic of Korea. Brain Tumor Res Treat. 2013; 1:16–23. PMID: 24904884.
Article10. Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010; 97:311–322. PMID: 19841865.
Article11. Lantos PL, Louis DN, Rosenblum Mk, Kleihues P. Tumors of the Nervous System. London: Oxford University Press;2002.12. Lee SH, Hwang TS, Koh YC, Kim WY, Han HS, Kim WS, et al. A consideration of MGMT gene promotor methylation analysis for glioblastoma using methylation-specific polymerase chain reaction and pyrosequencing. Korean J Pathol. 2011; 45:21–29.
Article13. Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007; 373:15–24. PMID: 17185754.14. Mikeska T, Bock C, El-Maarri O, Hübner A, Ehrentraut D, Schramm J, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007; 9:368–381. PMID: 17591937.
Article15. Park JW, Hwang JH. The long-term survival in patients with anaplastic astrocytoma or glioblastoma. J Korean Neurosurg Soc. 2003; 34:514–520.16. Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000; 60:5954–5958. PMID: 11085511.17. Provenzale JM, Ison C, Delong D. Bidimensional measurements in brain tumors : assessment of interobserver variability. AJR Am J Roentgenol. 2009; 193:W515–W522. PMID: 19933626.18. Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, et al. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990; 10:4987–4989. PMID: 1697035.
Article19. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study : 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009; 10:459–466. PMID: 19269895.
Article20. Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br Med Bull. 2008; 85:17–33. PMID: 18245773.
Article21. Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008; 10:332–337. PMID: 18556773.
Article22. Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997; 17:5612–5619. PMID: 9271436.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Consideration of MGMT Gene Promotor Methylation Analysis for Glioblastoma Using Methylation-Specific Polymerase Chain Reaction and Pyrosequencing
- Clinical and Prognostic Significance of Oâ¶-Methylguanine-DNA Methyltransferase Promoter Methylation in Patients with Melanoma: A Systematic Meta-Analysis
- Promoter hypermethylation of MGMT (O6-methylguanine-DNA methyltransferase) in cervical squamous cell carcinoma
- MGMT Gene Promoter Methylation Analysis by Pyrosequencing of Brain Tumour
- Study of CpG Methylation of the p16 and MGMT Promoter in Hepatocellular Carcinoma