Ann Clin Microbiol.
2014 Jun;17(2):50-57. 10.5145/ACM.2014.17.2.50.
Characterization of Salmonella spp. Clinical Isolates in Gyeongsangbuk-do Province, 2012 to 2013
- Affiliations
-
- 1Division of Microbiology, Gyeongsangbuk-do Provincial Government Institute of Health and Environment, Cheongwon, Korea.
- 2Division of Enteric Diseases, Center for Infectious Diseases, Korea National Institute of Health, Cheongwon, Korea. jun49@hanmail.net
- KMID: 1971094
- DOI: http://doi.org/10.5145/ACM.2014.17.2.50
Abstract
- BACKGROUND
Extended-spectrum cephalosporins and fluoroquinolones are important antimicrobials for treating invasive salmonellosis, and emerging resistance to these antimicrobials is of paramount concern.
METHODS
A total of 30 Salmonella spp. clinical isolates recovered in Gyeongsangbuk-do from 2012 to 2013 were characterized using antibiotic resistance profiles and pulsed-field gel electrophoresis (PFGE).
RESULTS
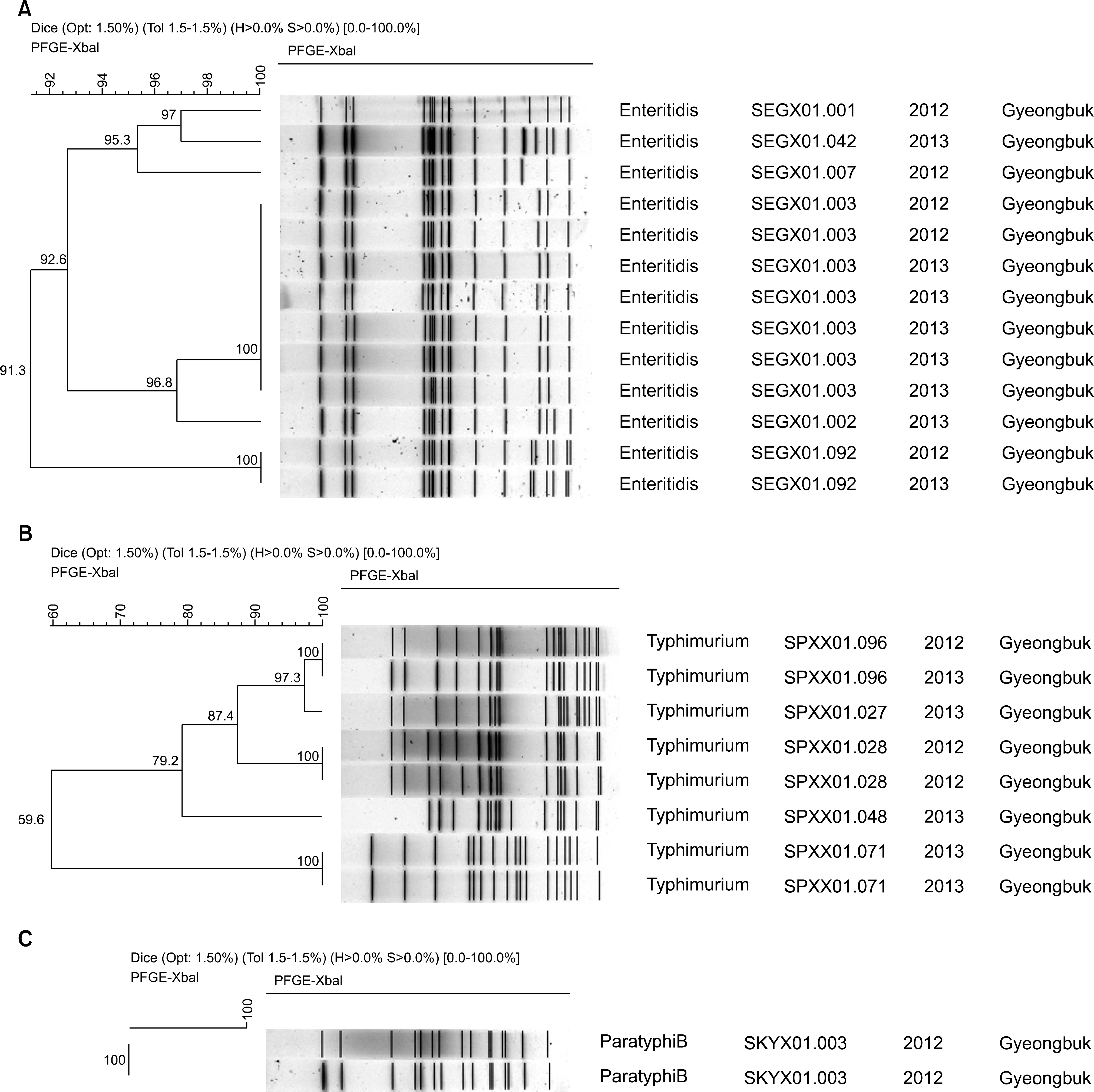
A high prevalence of multidrug-resistant isolates, mainly showing an ampicillin, nalidixic acid, chloramphenicol resistance pattern, was observed. Four extended-spectrum beta-lactamase (ESBL)-producing isolates (3 CTX-M-15 isolates and 1 CTX-M-27 isolate) were found. The bla(CTX-M-27) gene was carried by an IncF conjugative plasmid in the S. Infantis isolate. The bla(CTX-M-15) gene were carried by an IncF (2 isolates) or IncHI2 (1 isolate) conjugative plasmid in S. Enteritidis. In addition, a single mutation of GyrA, Ser83Thr (1 isolates), Asp87Tyr (9 isolates), Asp87Gly (4 isolates), and Asp87Leu (3 isolates), was detected in nalidixic acid-resistant Salmonella spp. isolates. XbaI PFGE analysis of all isolates revealed more than 19 different pulsotypes. The most common S. Enteritidis PFGE pattern (SEGX01.003) was associated with a larger number of cases of invasive salmonellosis than all other patterns.
CONCLUSION
The information from our study can assist in source attribution, outbreak investigations, and tailoring of interventions to maximize disease prevention.
Keyword
MeSH Terms
-
Ampicillin
beta-Lactamases
Cephalosporins
Chloramphenicol Resistance
Drug Resistance
Drug Resistance, Microbial
Electrophoresis, Gel, Pulsed-Field
Fluoroquinolones
Gyeongsangbuk-do
Nalidixic Acid
Plasmids
Prevalence
Salmonella Infections
Salmonella*
Ampicillin
Cephalosporins
Fluoroquinolones
Nalidixic Acid
beta-Lactamases
Figure
Reference
-
1.Khakhria R., Woodward D., Johnson WM., Poppe C. Salmonella isolated from humans, animals and other sources in Canada, 1983-92. Epidemiol Infect. 1997. 119:15–23.2.Jones TF., Ingram LA., Cieslak PR., Vugia DJ., Tobin-D'Angelo M., Hurd S, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008. 198:109–14.
Article3.Mermin J., Hutwagner L., Vugia D., Shallow S., Daily P., Bender J, et al. Emerging Infections Program FoodNet Working Group. Reptiles, amphibians, and human Salmonella infection: a population- based, case-control study. Clin Infect Dis. 2004. 38(Suppl 3):S253–61.4.Kimura AC., Reddy V., Marcus R., Cieslak PR., Mohle-Boetani JC., Kassenborg HD, et al. Emerging Infections Program FoodNet Working Group. Chicken consumption is a newly identified risk factor for sporadic Salmonella enterica serotype Enteritidis infections in the United States: a case-control study in FoodNet sites. Clin Infect Dis. 2004. 38(Suppl 3):S244–52.5.Scallan E., Hoekstra RM., Angulo FJ., Tauxe RV., Widdowson MA., Roy SL, et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011. 17:7–15.
Article6.Gatto AJ., Peters TM., Green J., Fisher IS., Gill ON., O'brien SJ, et al. Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000-2004: results of an international multi-centre study. Epidemiol Infect. 2006. 134:729–36.7.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001. 14:933–51.8.Oteo J., Aracil B., Alós JI., Gómez-Garcés JL. High rate of resistance to nalidixic acid in Salmonella enterica: its role as a marker of resistance to fluoroquinolones. Clin Microbiol Infect. 2000. 6:273–6.9.McEwen SA., Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002. 34(Suppl 3):S93–106.
Article10.Ewing WH. Edwards and Ewing's identification of Enterobacteriaceae. New York; Elsevier Science Publishing Co. Inc. 1986.11.CLSI. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. Document M100-S20. Wayne, PA; Clinical and Laboratory Standards Institute. 2010.12.Kim JY., Jeon SM., Rhie HG., Lee BK., Park MS., Lee H, et al. Rapid detection of extended spectrum β-lactamase (ESBL) for Enterobacteriaceae by use of a multiplex PCR-based method. Infect Chemother. 2009. 41:181–4.13.Kim JS., Kim J., Kim SJ., Jeon SE., Oh KH., Cho SH, et al. Characterization of CTX-M-type extended-spectrum beta-lactamase- producing diarrheagenic Escherichia coli isolates in the Republic of Korea during 2008-2011. J Microbiol Biotechnol. 2014. 24:421–6.14.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins KL., Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005. 63:219–28.
Article15.Casin I., Breuil J., Darchis JP., Guelpa C., Collatz E. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg Infect Dis. 2003. 9:1455–7.16.CDC. Standardized laboratory protocol for molecular subtyping of Escherchia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by Pulse-Field Gel Electrophoresis (PFGE). http://www.pulsenetinternational.org/assets/PulseNet/uploads/pfge/PNL05_Ec-Sal-ShigPFGEprotocol.pdf. [Online.] (last visited on 04 April 2014).17.Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001. 32:263–9.18.Biedenbach DJ., Toleman M., Walsh TR., Jones RN. Analysis of Salmonella spp. with resistance to extended-spectrum cephal-osporins and fluoroquinolones isolated in North America and Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997-2004). Diagn Microbiol Infect Dis. 2006. 54:13–21.19.KARMS. Korean Antimicrobial Resistance Monitoring System. KCDC. 2011.20.Colom K., Pérez J., Alonso R., Fernández-Aranguiz A., Lariño E., Cisterna R. Simple and reliable multiplex PCR assay for detection of bla TEM, bla SHV and bla OXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003. 223:147–51.21.Melano R., Corso A., Petroni A., Centrón D., Orman B., Pereyra A, et al. Multiple antibiotic-resistance mechanisms including a novel combination of extended-spectrum beta-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J Antimicrob Chemother. 2003. 52:36–42.22.Hopkins KL., Liebana E., Villa L., Batchelor M., Threlfall EJ., Carattoli A. Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob Agents Chemother. 2006. 50:3203–6.23.Tamang MD., Nam HM., Kim TS., Jang GC., Jung SC., Lim SK. Emergence of extended-spectrum beta-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J Clin Microbiol. 2011. 49:2671–5.24.Drlica K., Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997. 61:377–92.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence and antimicrobial resistance of Salmonella spp. and Escherichia coli isolated from ducks in Korea
- Prevalence of Salmonella spp. in pet turtles and their environment
- Application of SYBR Green real-time PCR assay for the specific detection of Salmonella spp
- Serovars distribution and antimicrobial resistance patterns of Salmonella spp. isolated from the swine farms and slaughter houses
- Usefulness of selenite F enrichment broth for the isolation of Salmonella from stool