Ann Lab Med.
2014 Jul;34(4):279-285. 10.3343/alm.2014.34.4.279.
Comparison of the Digene HPV Genotyping LQ Test and the PANArray HPV Genotyping Chip for Detection of High-Risk or Probable High-Risk Human Papillomavirus Genotypes
- Affiliations
-
- 1Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. changski@skku.edu
- KMID: 1791935
- DOI: http://doi.org/10.3343/alm.2014.34.4.279
Abstract
- BACKGROUND
We evaluated the performance of two different array-based techniques, a bead-based multiplex genotyping method (LQ; digene HPV Genotyping LQ Test, QIAGEN, Germany) and a DNA chip-based method using peptide nucleic acid probes (PANArray; PANArray HPV Genotyping Chip, Panagene, Korea), for detection of human papillomavirus (HPV) and genotyping of high-risk (HR) or probable high-risk (PHR) HPVs in healthy patients who visited a health-promotion center.
METHODS
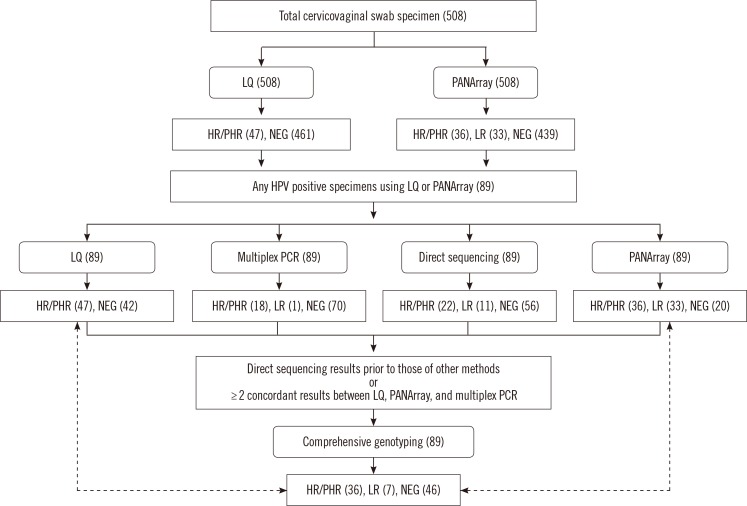
We obtained 508 unselected, consecutive cervicovaginal swab specimens. All specimens were examined by using the PANArray and LQ tests. All HPV-positive samples were then analyzed by multiplex PCR and direct sequencing.
RESULTS
The LQ test detected 47 HPV-positive cases (9.3%) with HR or PHR genotypes and the PANArray test identified 36 cases (7.1%). When the results of LQ and PANArray were compared by using comprehensive genotyping (integrated interpretation of the results of LQ, PANArray, multiplex PCR, and direct sequencing) for the detection of HR or PHR genotypes, the kappa values were 0.44 and 0.30 for LQ and PANArray, respectively. In comparison to comprehensive genotyping, the LQ test yielded 53 (60.0%) concordant and 12 (13.5%) compatible results, and the PANArray yielded 36 (40.4%) concordant and three (3.4%) compatible results.
CONCLUSIONS
The results of the LQ test had higher concordance and/or greater compatibility with those of comprehensive genotyping for the detection of HR or PHR genotypes than those of the PANArray test.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
Clinical Usefulness of a DNA Microarray-based Assay for the Diagnosis of Sexually Transmitted Infections
Ae Ja Park, So Young Kim, Dong Hee Seo
Lab Med Online. 2016;6(3):171-175. doi: 10.3343/lmo.2016.6.3.171.Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture II Assay in Cervicovaginal Swabs
Ho Young Jung, Hye Seung Han, Hyo Bin Kim, Seo Young Oh, Sun-Joo Lee, Wook Youn Kim
J Pathol Transl Med. 2016;50(2):138-146. doi: 10.4132/jptm.2015.10.21.
Reference
-
1. Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010; 401:70–79. PMID: 20206957.
Article2. Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003; 348:518–527. PMID: 12571259.
Article3. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009; 10:321–322. PMID: 19350698.
Article4. Poljak M, Kocjan BJ. Commercially available assays for multiplex detection of alpha human papillomaviruses. Expert Rev Anti Infect Ther. 2010; 8:1139–1162. PMID: 20954880.
Article5. Hwang Y, Lee M. Comparison of the AdvanSure human papillomavirus screening real-time PCR, the Abbott RealTime High Risk human papillomavirus test, and the Hybrid Capture human papillomavirus DNA test for the detection of human papillomavirus. Ann Lab Med. 2012; 32:201–205. PMID: 22563555.
Article6. Loy A, Bodrossy L. Highly parallel microbial diagnostics using oligonucleotide microarrays. Clin Chim Acta. 2006; 363:106–119. PMID: 16126187.
Article7. Clewley JP. A role for arrays in clinical virology: fact or fiction? J Clin Virol. 2004; 29:2–12. PMID: 14675863.
Article8. Godínez JM, Tous S, Baixeras N, Moreno-Crespi J, Alejo M, Lejeune M, et al. Performance of the digene LQ, RH and PS HPVs genotyping systems on clinical samples and comparison with HC2 and PCR-based Linear Array. Infect Agent Cancer. 2011; 6:23. PMID: 22093164.
Article9. Geraets DT, Lenselink CH, Bekkers RL, van Doorn LJ, Quint WG, Melchers WJ. Universal human papillomavirus genotyping by the digene HPV Genotyping RH and LQ Tests. J Clin Virol. 2011; 50:276–280. PMID: 21296612.
Article10. Zubach V, Smart G, Ratnam S, Severini A. Novel microsphere-based method for detection and typing of 46 mucosal human papillomavirus types. J Clin Microbiol. 2012; 50:460–464. PMID: 22116162.
Article11. Cho EJ, Do JH, Kim YS, Bae S, Ahn WS. Evaluation of a liquid bead array system for high-risk human papillomavirus detection and genotyping in comparison with Hybrid Capture II, DNA chip and sequencing methods. J Med Microbiol. 2011; 60:162–171. PMID: 20965919.
Article12. Choi JJ, Kim C, Park H. Peptide nucleic acid-based array for detecting and genotyping human papillomaviruses. J Clin Microbiol. 2009; 47:1785–1790. PMID: 19369432.
Article13. Husnjak K, Grce M, Magdić L, Pavelić K. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J Virol Methods. 2000; 88:125–134. PMID: 10960700.
Article14. Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlée F, Hildesheim A, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000; 38:357–361. PMID: 10618116.
Article15. Nishiwaki M, Yamamoto T, Tone S, Murai T, Ohkawara T, Matsunami T, et al. Genotyping of human papillomaviruses by a novel one-step typing method with multiplex PCR and clinical applications. J Clin Microbiol. 2008; 46:1161–1168. PMID: 18234872.
Article16. Geraets DT, Heideman DA, de Koning MN, Snijders PJ, van Alewijk DC, Meijer CJ, et al. High-throughput genotyping of high-risk HPV by the digene HPV Genotyping LQ Test using GP5+/6+-PCR and xMAP technology. J Clin Virol. 2009; 46(Suppl 3):S21–S26. PMID: 20129070.
Article17. Lee JK, Kim MK, Song SH, Hong JH, Min KJ, Kim JH, et al. Comparison of human papillomavirus detection and typing by hybrid capture 2, linear array, DNA chip, and cycle sequencing in cervical swab samples. Int J Gynecol Cancer. 2009; 19:266–272. PMID: 19396007.
Article18. Song SH, Hong JH, Kwak SH, Lee JK, Kim MK. Clinical performance assessment of five human papillomavirus DNA tests using liquid-based cytology samples. J Obstet Gynaecol Res. 2012; 38:408–414. PMID: 22175246.
Article19. Chung MY, Kim YW, Bae SM, Kwon EH, Chaturvedi PK, Battogtokh G, et al. Development of a bead-based multiplex genotyping method for diagnostic characterization of HPV infection. PLoS One. 2012; 7:e32259. PMID: 22393393.
Article20. Schmitz M, Scheungraber C, Herrmann J, Teller K, Gajda M, Runnebaum IB, et al. Quantitative multiplex PCR assay for the detection of the seven clinically most relevant high-risk HPV types. J Clin Virol. 2009; 44:302–307. PMID: 19223232.
Article21. Carcopino X, Henry M, Mancini J, Giusiano S, Boubli L, Olive D, et al. Significance of HPV 16 and 18 viral load quantitation in women referred for colposcopy. J Med Virol. 2012; 84:306–313. PMID: 22170552.
Article22. Gharizadeh B, Oggionni M, Zheng B, Akom E, Pourmand N, Ahmadian A, et al. Type-specific multiple sequencing primers: a novel strategy for reliable and rapid genotyping of human papillomaviruses by pyrosequencing technology. J Mol Diagn. 2005; 7:198–205. PMID: 15858143.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparative Evaluation of the HPV28 Detection and HPV DNA Chip Test for Detecting and Genotyping Human Papillomaviruses
- Comparison of Four Human Papillomavirus Genotyping Methods: Next-generation Sequencing, INNO-LiPA, Electrochemical DNA Chip, and Nested-PCR
- Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture II Assay in Cervicovaginal Swabs
- Evaluation of clinical usefulness of HPV-16 and HPV-18 genotyping for cervical cancer screening
- Clinical significance of human papillomavirus genotyping