Ann Lab Med.
2018 Mar;38(2):139-146. 10.3343/alm.2018.38.2.139.
Comparison of Four Human Papillomavirus Genotyping Methods: Next-generation Sequencing, INNO-LiPA, Electrochemical DNA Chip, and Nested-PCR
- Affiliations
-
- 1Center of Excellence in Clinical Virology, Department of Pediatrics, Faculty of Medicine, Chulalongkorn University, Bangkok. yong.p@chula.ac.th
- 2Research Affairs, Faculty of Medicine, Chulalongkorn University, Bangkok.
- 3Department of Biochemistry, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
- KMID: 2403359
- DOI: http://doi.org/10.3343/alm.2018.38.2.139
Abstract
- BACKGROUND
Human papillomavirus (HPV) infection causes cervical cancer, thus necessitating early detection by screening. Rapid and accurate HPV genotyping is crucial both for the assessment of patients with HPV infection and for surveillance studies.
METHODS
Fifty-eight cervicovaginal samples were tested for HPV genotypes using four methods in parallel: nested-PCR followed by conventional sequencing, INNO-LiPA, electrochemical DNA chip, and next-generation sequencing (NGS).
RESULTS
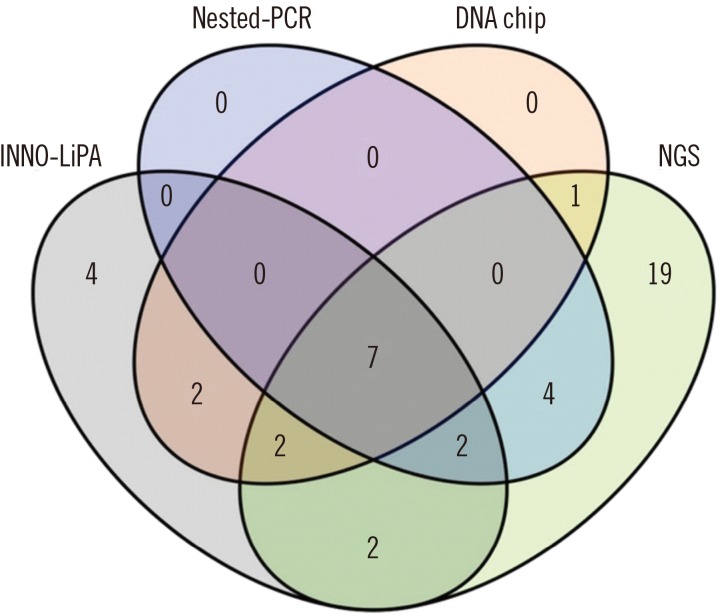
Seven HPV genotypes (16, 18, 31, 33, 45, 56, and 58) were identified by all four methods. Nineteen HPV genotypes were detected by NGS, but not by nested-PCR, INNO-LiPA, or electrochemical DNA chip.
CONCLUSIONS
Although NGS is relatively expensive and complex, it may serve as a sensitive HPV genotyping method. Because of its highly sensitive detection of multiple HPV genotypes, NGS may serve as an alternative for diagnostic HPV genotyping in certain situations.
Keyword
MeSH Terms
Figure
Reference
-
1. Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011; 22:2675–2686. PMID: 21471563.2. Allan B, Marais DJ, Hoffman M, Shapiro S, Williamson AL. Cervical human papillomavirus (HPV) infection in South African women: implications for HPV screening and vaccine strategies. J Clin Microbiol. 2008; 46:740–742. PMID: 17977997.3. Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008; 26:K1–K16. PMID: 18847553.4. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004; 324:17–27. PMID: 15183049.5. Chen L, Watanabe K, Haruyama T, Kobayashi N. Simple and rapid human papillomavirus genotyping method by restriction fragment length polymorphism analysis with two restriction enzymes. J Med Virol. 2013; 85:1229–1234. PMID: 23918541.6. de Antonio JC, Fernández-Olmos A, Mercadillo M, Lindemann ML, Mochales FB. Detection of high-risk human papillomavirus by two molecular techniques: hybrid capture and linear array. J Virol Methods. 2008; 149:163–166. PMID: 18328575.7. Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinsky SM. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989; 7:209–214.8. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995; 76:1057–1062. PMID: 9049358.9. Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, et al. Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. J Virol Methods. 2004; 122:87–93. PMID: 15488625.10. van Hamont D, van Ham MA, Bakkers JM, Massuger LF, Melchers WJ. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the roche linear array HPV genotyping test. J Clin Microbiol. 2006; 44:3122–3129. PMID: 16954236.11. Coutlée F, Rouleau D, Petignat P, Ghattas G, Kornegay JR, Schlag P, et al. Enhanced detection and typing of human papillomavirus (HPV) DNA in anogenital samples with PGMY primers and the Linear array HPV genotyping test. J Clin Microbiol. 2006; 44:1998–2006. PMID: 16757590.12. Chansaenroj J, Theamboonlers A, Chinchai T, Junyangdikul P, Swangvaree S, Karalak A, et al. High-risk human papillomavirus genotype detection by electrochemical DNA chip method. Asian Pac J Cancer Prev. 2012; 13:1151–1158. PMID: 22799297.13. Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next-generation sequencing technologies in virology. J Gen Virol. 2012; 93:1853–1868. PMID: 22647373.14. Sun C, McAndrew T, Smith BC, Chen Z, Frimer M, Burk RD. Characterization of HPV DNA methylation of contiguous CpG sites by bisulfite treatment and massively parallel sequencing-the FRAGMENT approach. Front Genet. 2014; 5:150. PMID: 24917876.15. Yi X, Zou J, Xu J, Liu T, Liu T, Hua S, et al. Development and validation of a new HPV genotyping assay based on next-generation sequencing. Am J Clin Pathol. 2014; 141:796–804. PMID: 24838323.16. Chinchai T, Chansaenroj J, Junyangdikul P, Swangvaree S, Karalak A, Niruthisard S, et al. Comparison between direct sequencing and INNO-LiPA methods for HPV detection and genotyping in Thai Women. Asian Pac J Cancer Prev. 2011; 12:989–994. PMID: 21790239.17. Lurchachaiwong W, Junyangdikul P, Payungporn S, Chansaenroj J, Sampatanukul P, Tresukosol D, et al. Relationship between hybrid capture II ratios and DNA amplification of E1, E6 and L1 genes used for the detection of human papillomavirus in samples with different cytological findings. Asian Pac J Allergy Immunol. 2009; 27:217–224. PMID: 20232576.18. Ermel A, Qadadri B, Morishita A, Miyagawa I, Yamazaki G, Weaver B, et al. Human papillomavirus detection and typing in thin prep cervical cytologic specimens comparing the Digene Hybrid Capture II Assay, the Roche Linear Array HPV Genotyping Assay, and the Kurabo GeneSquare Microarray Assay. J Virol Methods. 2010; 169:154–161. PMID: 20670658.19. Barzon L, Militello V, Lavezzo E, Franchin E, Peta E, Squarzon L, et al. Human papillomavirus genotyping by 454 next generation sequencing technology. J Clin Virol. 2011; 52:93–97. PMID: 21802982.20. Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008; 5:16–18. PMID: 18165802.21. Smith BC, McAndrew T, Chen Z, Harari A, Barris DM, Viswanathan S, et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLoS One. 2012; 7:e40425. PMID: 22792313.22. Cheng YP, Chen CW, Sheen YS, Tsai TF. Genotype distribution of human papillomavirus in anogenital warts of male patients in Taiwan. Derm Sinica. 2012; 30:85–89.23. Menzo S, Monachetti A, Trozzi C, Ciavattini A, Carloni G, Varaldo PE, et al. Identification of six putative novel human papillomaviruses (HPV) and characterization of candidate HPV type 87. J Virol. 2001; 75:11913–11919. PMID: 11689676.24. Maver PJ, Kocjan BJ, Seme K, Poljak M. Genomic diversity of low-risk human papillomavirus genotypes HPV 40, HPV 42, HPV 43, and HPV 44. J Med Virol. 2014; 86:272–282. PMID: 24155245.25. Sasagawa T, Basha W, Yamazaki H, Inoue M. High-risk and multiple human papillomavirus infections associated with cervical abnormalities in Japanese women. Cancer Epidemiol Biomarkers Prev. 2001; 10:45–52. PMID: 11205488.26. International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. Human papillomaviruses, volume 90. Lyon, France: International Agency for Research on Cancer;2007.27. Muñoz N, Castellsagué X, de González AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006; 24:S3/1-10.28. Micalessi MI, Boulet GA, Bogers J. A real-time PCR approach based on SPF10 primers and the INNO-LiPA HPV genotyping extra assay for the detection and typing of human papillomavirus. Methods Mol Biol. 2015; 1249:27–35. PMID: 25348295.29. Perrons C, Kleter B, Jelley R, Jalal H, Quint W, Tedder R. Detection and genotyping of human papillomavirus DNA by SPF10 and MY09/11 primers in cervical cells taken from women attending a colposcopy clinic. J Med Virol. 2002; 67:246–252. PMID: 11992586.30. da Fonseca AJ, Galvão RS, Miranda AE, Ferreira LC, Chen Z. Comparison of three human papillomavirus DNA detection methods: next generation sequencing, multiplex-PCR and nested-PCR followed by Sanger based sequencing. J Med Virol. 2016; 88:888–894. PMID: 26496186.31. Abreu AL, Souza RP, Gimenes F, Consolaro ME. A review of methods for detect human Papillomavirus infection. Virol J. 2012; 9:262. PMID: 23131123.32. Salazar KL, Zhou HS, Xu J, Peterson LE, Schwartz MR, Mody DR, et al. Multiple human papilloma virus infections and their impact on the development of high-risk cervical lesions. Acta Cytol. 2015; 59:391–398. PMID: 26674365.33. Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlée F, Hildesheim A, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000; 38:357–361. PMID: 10618116.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of the AdvanSure Human Papillomavirus Screening Real-Time PCR, the Abbott RealTime High Risk Human Papillomavirus Test, and the Hybrid Capture Human Papillomavirus DNA Test for the Detection of Human Papillomavirus
- Comparison of the Digene HPV Genotyping LQ Test and the PANArray HPV Genotyping Chip for Detection of High-Risk or Probable High-Risk Human Papillomavirus Genotypes
- Comparative Evaluation of the HPV28 Detection and HPV DNA Chip Test for Detecting and Genotyping Human Papillomaviruses
- Genotyping of Human Papillomavirus Detected with the HPV DNA Chip in Cervical Cancer
- A Comparative Study of Apolipoprotein E Genotyping: INNO-LiPA ApoE Kit, Allelic Discrimination with LightCycler, and BioCore ApoE Kit