Ann Lab Med.
2014 Mar;34(2):111-117. 10.3343/alm.2014.34.2.111.
Performance of Kiestra Total Laboratory Automation Combined with MS in Clinical Microbiology Practice
- Affiliations
-
- 1Department of Infectious Diseases, Medical Microbiology and Hygiene, Heidelberg University Hospital, Heidelberg, Germany. nico.mutters@med.uni-heidelberg.de
- 2Academic Medical Centre, Department of Medical Microbiology, University of Amsterdam, Amsterdam, the Netherlands.
- KMID: 1791903
- DOI: http://doi.org/10.3343/alm.2014.34.2.111
Abstract
- BACKGROUND
Microbiological laboratories seek technologically innovative solutions to cope with large numbers of samples and limited personnel and financial resources. One platform that has recently become available is the Kiestra Total Laboratory Automation (TLA) system (BD Kiestra B.V., the Netherlands). This fully automated sample processing system, equipped with digital imaging technology, allows superior detection of microbial growth. Combining this approach with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MS) (Bruker Daltonik, Germany) is expected to enable more rapid identification of pathogens.
METHODS
Early growth detection by digital imaging using Kiestra TLA combined with MS was compared to conventional methods (CM) of detection. Accuracy and time taken for microbial identification were evaluated for the two methods in 219 clinical blood culture isolates. The possible clinical impact of earlier microbial identification was assessed according to antibiotic treatment prescription.
RESULTS
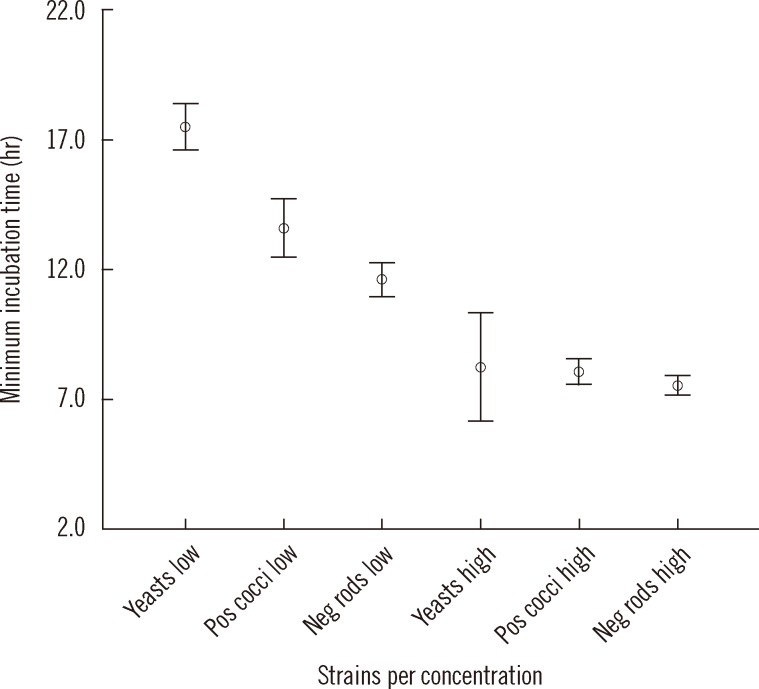
Pathogen identification using Kiestra TLA combined with MS resulted in a 30.6 hr time gain per isolate compared to CM. Pathogens were successfully identified in 98.4% (249/253) of all tested isolates. Early microbial identification without susceptibility testing led to an adjustment of antibiotic regimen in 12% (24/200) of patients.
CONCLUSIONS
The requisite 24 hr incubation time for microbial pathogens to reach sufficient growth for susceptibility testing and identification would be shortened by the implementation of Kiestra TLA in combination with MS, compared to the use of CM. Not only can this method optimize workflow and reduce costs, but it can allow potentially life-saving switches in antibiotic regimen to be initiated sooner.
Keyword
MeSH Terms
-
Automation, Laboratory
Candida albicans/genetics/*isolation & purification
Disk Diffusion Antimicrobial Tests
Gram-Negative Bacteria/genetics/*isolation & purification
Gram-Positive Bacteria/genetics/*isolation & purification
Humans
RNA, Ribosomal, 16S/chemistry/genetics
Retrospective Studies
Sequence Analysis, RNA
*Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization
RNA, Ribosomal, 16S
Figure
Reference
-
1. Carroll KC, Weinstein MP. Manual and automated systems for detection and identification of microorganisms. In : Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of clinical microbiology. 9th ed. Washington, DC: American Society for Microbiology;2007. p. 192–217.2. Greub G, Prod'hom G. Automation in clinical bacteriology: what system to choose? Clin Microbiol Infect. 2011; 17:655–660. PMID: 21521409.
Article3. Bourbeau PP, Ledeboer NA. Automation in clinical microbiology. J Clin Microbiol. 2013; 51:1658–1665. PMID: 23515547.
Article4. Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis. 2013; 57:1237–1245. PMID: 23899684.
Article5. Dermoumi H. Fungi as pathogens in infection--results of a multicenter study. Mykosen. 1987; 30:349–354. PMID: 3313037.6. Foltzer MA, Reese RE. Bacteremia and sepsis. In : Reese RE, Betts RF, editors. A practical approach to infectious diseases. 3rd ed. Boston: Little Brown and Company;1991. p. 19–53.7. Grosse-Herrenthey A, Maier T, Gessler F, Schaumann R, Böhnel H, Kostrzewa M, et al. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe. 2008; 14:242–249. PMID: 18621134.
Article8. Drancourt M, Berger P, Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004; 42:2197–2202. PMID: 15131188.
Article9. van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010; 48:900–907. PMID: 20053859.
Article10. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009; 49:543–551. PMID: 19583519.
Article11. Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010; 48:1169–1175. PMID: 20164271.
Article12. Eigner U, Holfelder M, Oberdorfer K, Betz-Wild U, Bertsch D, Fahr AM. Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin Lab. 2009; 55:289–296. PMID: 19894408.14. Matthews S, Deutekom J. The future of diagnostic bacteriology. Clin Microbiol Infect. 2011; 17:651–654. PMID: 21521408.
Article15. La Scola B, Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009; 4:e8041. PMID: 19946369.
Article16. Klein S, Zimmermann S, Köehler C, Mischnik A, Alle W, Bode K. Integration of matrix-assisted laser desorpotion/ionization time-of-flight mass spectrometry in bloodculture diagnostic:a fast and effective approach. J Med Microbiol. 2012; 61:323–331. PMID: 22074848.17. Loonen AJ, Jansz AR, Stalpers J, Wolffs PF, van den Brule AJ. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2012; 31:1575–1583. PMID: 22080416.
Article18. Madhavan P, Jamal F, Chong PP, Ng KP. Identification of local clinical Candida isolates using CHROMagar Candida™ as a primary identification method for various Candida species. Trop Biomed. 2011; 28:269–274. PMID: 22041745.19. Gherardi G, Angeletti S, Panitti M, Pompilio A, Di Bonaventura G, Crea F, et al. Comparative evaluation of the Vitek-2 Compact and Phoenix systems for rapid identification andantibiotic susceptibility testing directly from blood cultures of Gram-negative and Gram-positive isolates. Diagn Microbiol Infect Dis. 2012; 72:20–31. PMID: 22030102.20. Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med. 2003; 115:529–535. PMID: 14599631.
Article21. Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005; 365:63–78. PMID: 15639681.
Article22. Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005; 49:760–766. PMID: 15673761.
Article23. Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob Agents Chemother. 2005; 49:1306–1311. PMID: 15793102.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of the Performance of an Adiponectin ELISA-based Test and Establishing Serum Adiponectin Reference Intervals for Korean Population
- Evaluation of automated calibration and quality control processes using the Aptio total laboratory automation system
- Performance Evaluation of the DxH 800 Hematology Analyzer
- Performance Evaluation of Automated Chemistry Analyser for Urine Chemistry Test
- Comparison of Red Blood Cell, White Blood Cell and Differential Counts between UF-5000 System and Manual Method