Korean J Lab Med.
2010 Aug;30(4):381-387. 10.3343/kjlm.2010.30.4.381.
A Comparison Analysis on the Diagnosis of Helicobacter pylori Infection and the Detection of Clarithromycin Resistance according to Biopsy Sites
- Affiliations
-
- 1Department of Laboratory Medicine, Chung-Ang University College of Medicine, Seoul, Korea. cpworld@cau.ac.kr
- KMID: 1781644
- DOI: http://doi.org/10.3343/kjlm.2010.30.4.381
Abstract
- BACKGROUND
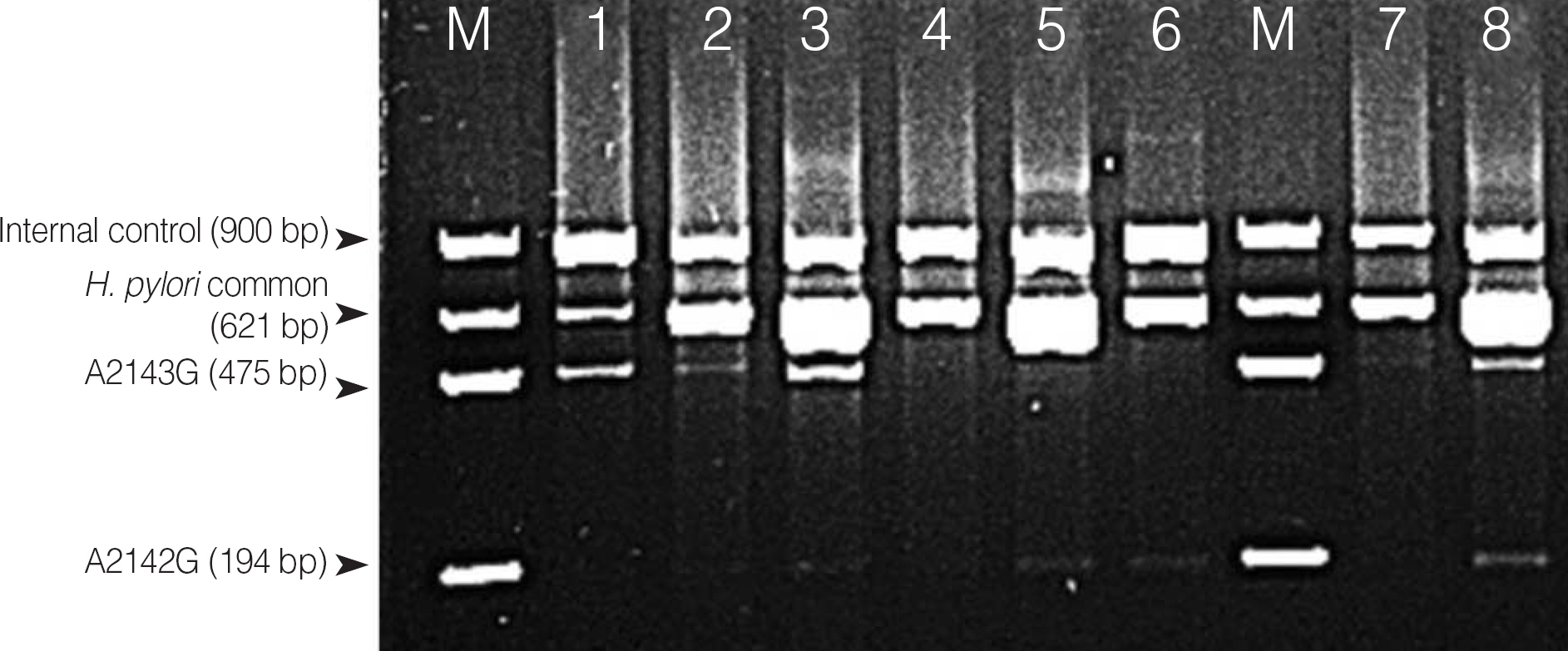
This study was performed to determine the biopsy sites that are suitable for the diagnosis of Helicobacter pylori infection and to assess the sensitivity of culture, histology, and dual-priming oligonucleotide (DPO)-based multiplex PCR. Moreover, we evaluated the usefulness of PCR for the detection of 23S rRNA mutations, which are responsible for the clarithromycin resistance of H. pylori.
METHODS
From 90 patients, we obtained biopsy specimens for culture, histology, and Seeplex(R) ClaR-H. pylori PCR (Seegene Inc., Korea). Phenotypic susceptibility to clarithromycin was evaluated using the E-test (AB Biodisk, Sweden).
RESULTS
H. pylori was detected in 48 of 90 patients. The positive rates of infection in the antrum and body were higher than those in the biopsies obtained from the duodenal bulb. The positive rates in histology, PCR, and culture were 46.7%, 42.2%, and 34.4%, respectively. Using histology or PCR, we identified H. pylori in 46 of the 48 patients. 23S rRNA mutations were detected in 8 patients. The clarithromycin E-test showed that all the 10 wild-type patients were susceptible. However, the results of the PCR and E-test of 3 of the 8 mutation-positive patients were discrepant.
CONCLUSIONS
We observed that a combination of histology and PCR affords a high detection rate of H.pylori infection and that DPO-based PCR can be practically used for the diagnosis of H. pylori infection and the determination of clarithromycin resistance. These techniques were useful for biopsy sampling simultaneously from the antrum and body for the detection of clarithromycin resistance of multiple strain infection or heteroresistance.
MeSH Terms
-
Anti-Bacterial Agents/*pharmacology
Biopsy
Clarithromycin/*pharmacology
Drug Resistance, Bacterial
Genotype
Helicobacter Infections/*diagnosis/drug therapy/pathology
Helicobacter pylori/drug effects/genetics/*isolation & purification
Humans
Microbial Sensitivity Tests
Mutation
Polymerase Chain Reaction
RNA, Ribosomal, 23S/genetics
Figure
Reference
-
1.NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA. 1994. 272:65–9.2.Blaser MJ. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999. 179:1523–30.3.Pounder RE., Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995. 9(S2):33–9.4.Yim JY., Kim N., Choi SH., Kim YS., Cho KR., Kim SS, et al. Seroprevalence of Helicobacter pylori in South Korea. Helicobacter. 2007. 12:333–40.5.Lee MS., Roe IH., Lee MY., Lee JH., Nam SW., Lee MI, et al. The best endoscopic biops site for the diagnosis of Helicobacter pylori infection. Korean J Gastroenterol. 2001. 37:406–11. (이문숙, 노임환, 이만용, 이재현, 남승우, 이명인 등. Helicobacter pylori 감염을 진단하기위한 내시경 생검의 최적 위치. 대한소화기학회지 2001;37:406-11.).6.Kim N., Kim JJ., Choe YH., Kim HS., Kim JI., Chung IS. Diagnosis and treatment guidelines for Helicobacter pylori infection in Korea. Korean J Gastroenterol. 2009. 54:269–78. (김나영, 김재준, 최연호, 김현수,김진일, 정인식. 헬리코박터 파일로리 감염의 진단 및 치료 가이드라인. Korean J Gastroenterol 2009;54:269-78.).7.Malfertheiner P., Meégraud F., O'Morain C., Hungin AP., Jones R., Axon A, et al. Current concepts in the management of Helicobacter pylori infection—the Maastricht 2-2000 Consensus Report. Aliment Pharmacol Ther. 2002. 16:167–80.8.Lee HK., Chae HS., Kang JO., Lee MK., Sung H., Kim MN, et al. Multicenterstudy for the frequency of 23S rRNA point mutations associated with clarithromycin resistance in Helicobacter pylori in Korea. Korean J Clin Microbiol. 2008. 11:84–9. (이혜경, 채현석, 강정옥, 이미경, 성흥섭, 김미나 등. 한국에서 Helicobacter pylori 균주의 Clarithromycin 내성 23S rRNA 유전자 점돌연변이의 빈도에 대한 다기관 연구. Korean J Clin Microbiol 2008;11:84-9.).9.Kim JM. Antibiotic resistance of Helicobacter pylori isolated from Korean patients. Korean J Gastroenterol. 2006. 47:337–49. (김정목. 한 국인 환자에서 분리한 Helicobacter pylori 균주의 항생제 내성률. Korean J Gastroenterol 2006;47:33749.).10.Kim JM., Kim JS., Kim N., Kim SG., Jung HC., Song IS. Comparison of primary and secondary antimicrobial minimum inhibitory concentrations for Helicobacter pylori isolated from Korean patients. Int J Antimicrob Agents. 2006. 28:6–13.11.Sung H., Chung HJ., Kim MN., Lee GH. Clinical usefulness of antimicrobial susceptibility test for Helicobacter pylori. Korean J Lab Med. 2006. 26:179–84. (성흥섭, 정희정, 김미나, 이진혁. Helicobacter pylori 항균제 감수성 검사의 임상적 유용성. 대한진단검사의학회지 2006;26:179-84.).12.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Eighteenth informational supplement, M100-S18. Wayne, PA: CLSI;2008.13.Woo HY., Park DI., Park H., Kim MK., Kim DH., Kim IS, et al. Dual-priming oligonucleotide-based multiplex PCR for the detection of Helicobacter pylori and determination of clarithromycin resistance with gastric biopsy specimens. Helicobacter. 2009. 14:22–8.14.Byun YH., Jo YJ., Kim SC., Lee JS., Shin WY., Park YS, et al. Clinical factors that predicts successful eradication of Helicobacter pylori. Korean J Gastroenterol. 2006. 48:172–9. (변영혜, 조윤주, 김성철, 이준석,신원용, 박영숙 등Helicobacter pylori 감염 환자에서 제균을 예측할수 있는 임상 인자. Korean J Gastroenterol 2006;48:172-9.).15.Versalovic J., Osato MS., Spakovsky K., Dore MP., Reddy R., Stone GG, et al. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997. 40:283–6.16.Fontana C., Favaro M., Pietroiusti A., Pistoia ES., Galante A., Favalli C. Detection of clarithromycin-resistant Helicobacter pylori in stool samples. J Clin Microbiol. 2003. 41:3636–40.17.Gibson JR., Saunders NA., Burke B., Owen RJ. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999. 37:3746–8.18.Oleastro M., Menard A., Santos A., Lamouliatte H., Monteiro L., Barthélémy P, et al. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003. 41:397–402.19.Trebesius K., Panthel K., Strobel S., Vogt K., Faller G., Kirchner T, et al. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut. 2000. 46:608–14.20.Taylor DE., Ge Z., Purych D., Lo T., Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997. 41:2621–8.21.Kim JW., Kim JG., Chae SL., Cha YJ., Park SM. High prevalence of multiple strain colonization of Helicobacter pylori in Korean patients: DNA diversity among clinical isolates from the gastric corpus, antrum and duodenum. Korean J Intern Med. 2004. 19:1–9.22.Nam SW., Roe IH., Kim SB., Lee BS., Hwang YJ., Park HJ, et al. Detection of clarithromycin-resistant Helicobacter pylori by polymerase chain reaction. Korean J Gastroenterol. 2000. 36:450–6. (남승우, 노임환, 김석배, 이병석, 황영준, 박현종 등. Clarithromycin 내성을 지닌Helicobacter pylori 균주 확인을 위한 중합효소연쇄반응법의 이용. 대한소화기학회지 2000;36:450-6.).23.De Francesco V., Margiotta M., Zullo A., Hassan C., Troiani L., Burattini O, et al. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006. 144:94–100.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Eradication Therapy for Helicobacter pylori with Diagnostic Test for Clarithromycin Resistance
- Detection of 23S rRNA Mutation Associated with Clarithromycin Resistance in Children with Helicobacter pylori Infection
- Management of Helicobacter pylori Infection in Europe: Focusing on the Maastricht V/Florence Consensus
- Antimicrobial Resistance Rates in Helicobacter pylori and Detection of 23S rRNA Mutation Associated with Clarithromycin Resistance
- Patterns of Helicobacter pylori Resistance to Metronidazole, Clarithormycin and Amoxicillin in Saudi Arabia