J Korean Med Sci.
2009 Jun;24(3):493-497. 10.3346/jkms.2009.24.3.493.
Differential Methylation Pattern of ID4, SFRP1, and SHP1 between Acute Myeloid Leukemia and Chronic Myeloid Leukemia
- Affiliations
-
- 1Institute of Human Genetics, Department of Anatomy, Brain Korea 21 Biomedical Sciences, Korea University College of Medicine, Seoul, Korea. parksh@korea.ac.kr
- 2Division of Hematology/Oncology, Department of Internal Medicine, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea.
- KMID: 1779169
- DOI: http://doi.org/10.3346/jkms.2009.24.3.493
Abstract
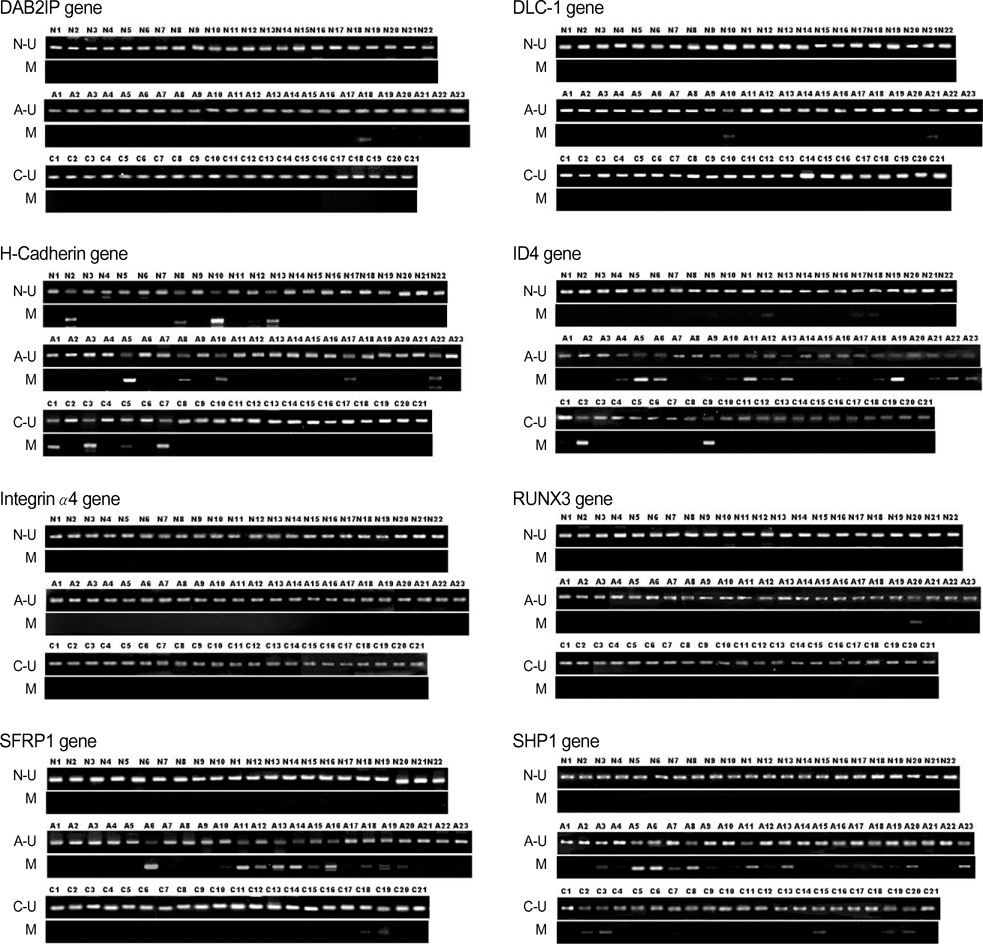
- To gain insight into the differential mechanism of gene promoter hypermethylation in acute and chronic leukemia, we identified the methylation status on one part of 5'CpG rich region of 8 genes, DAB2IP, DLC-1, H-cadherin, ID4, Integrin alpha4, RUNX3, SFRP1, and SHP1 in bone marrows from acute myeloid leukemia (AML) and chronic myeloid leukemia (CML) patients. Also, we compared the methylation status of genes in AML and CML using methylation-specific PCR (MSP). The frequencies of DNA methylation of ID4, SFRP1, and SHP1 were higher in AML patients compared to those in CML patients. In contrast, no statistical difference between AML and CML was detected for other genes such as DLC-1, DAB2IP, H-cadherin, Integrin alpha4, and RUNX3. Taken together, these results suggest that these methylation-controlled genes may have different roles in AML and CML, and thus, may act as a biological marker of AML.
MeSH Terms
-
Adolescent
Adult
Aged
CpG Islands
*DNA Methylation
Female
Humans
Inhibitor of Differentiation Proteins/*genetics/metabolism
Intercellular Signaling Peptides and Proteins/*genetics/metabolism
Leukemia, Myelogenous, Chronic, BCR-ABL Positive/genetics/metabolism
Leukemia, Myeloid, Acute/*genetics/metabolism
Male
Membrane Proteins/*genetics/metabolism
Middle Aged
Promoter Regions, Genetic
Protein Tyrosine Phosphatase, Non-Receptor Type 6/*genetics/metabolism
Figure
Reference
-
1. Wahlfors J, Hiltunen H, Heinonen K, Hamalainen E, Alhonen L, Janne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992. 80:2074–2080.
Article2. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002. 21:5400–5413.
Article3. Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, Jones PA, Lubbert M. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2'-deoxycytidine (decitabine) treatment. Blood. 2002. 100:2957–2964.
Article4. Fulop Z, Csernus B, Timar B, Szepesi A, Matolcsy A. Microsatellite instability and hMLH1 promoter hypermethylation in Richter's transformation of chronic lymphocytic leukemia. Leukemia. 2003. 17:411–415.
Article5. Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997. 272:22322–22329.6. Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005. 97:1498–1506.
Article7. Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, Baylin SB. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997. 57:837–841.8. Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999. 93:4347–4353.
Article9. Melki JR, Vincent PC, Brown RD, Clark SJ. Hypermethylation of E-cadherin in leukemia. Blood. 2000. 95:3208–3213.
Article10. Ng MH, Chung YF, Lo KW, Wickham NW, Lee JC, Huang DP. Frequent hypermethylation of p16 and p15 genes in multiple myeloma. Blood. 1997. 89:2500–2506.
Article11. Yano M, Toyooka S, Tsukuda K, Dote H, Ouchida M, Hanabata T, Aoe M, Date H, Gazdar AF, Shimizu N. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005. 113:59–66.12. Kim TY, Jong HS, Song SH, Dimtchev A, Jeong SJ, Lee JW, Kim NK, Jung M, Bang YJ. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003. 22:3943–3951.
Article13. Sun D, Zhang Z, Van do N, Huang G, Ernberg I, Hu L. Aberrant methylation of CDH13 gene in nasopharyngeal carcinoma could serve as a potential diagnostic biomarker. Oral Oncol. 2007. 43:82–87.
Article14. Chan AS, Tsui WY, Chen X, Chu KM, Chan TL, Chan AS, Li R, So S, Yuen ST, Leung SY. Downregulation of ID4 by promoter hypermethylation in gastric adenocarcinoma. Oncogene. 2003. 22:6946–6953.
Article15. Park J, Song SH, Kim TY, Choi MC, Jong HS, Lee JW, Kim NK, Kim WH, Bang YJ. Aberrant methylation of integrin alpha4 gene in human gastric cancer cells. Oncogene. 2004. 23:3474–3480.16. Long C, Yin B, Lu Q, Zhou X, Hu J, Yang Y, Yu F, Yuan Y. Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest. 2007. 25:685–690.
Article17. Takada T, Yagi Y, Maekita T, Imura M, Nakagawa S, Tsao SW, Miyamoto K, Yoshino O, Yasugi T, Taketani Y, Ushijima T. Methylation-associated silencing of the Wnt antagonist SFRP1 gene in human ovarian cancers. Cancer Sci. 2004. 95:741–744.
Article18. Khoury JD, Rassidakis GZ, Medeiros LJ, Amin HM, Lai R. Methylation of SHP1 gene and loss of SHP1 protein expression are frequent in systemic anaplastic large cell lymphoma. Blood. 2004. 104:1580–1581.
Article19. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996. 93:9821–9826.
Article20. Kim DG, Park SY, You KR, Lee GB, Kim H, Moon WS, Chun YH, Park SH. Establishment and characterization of chromosomal aberrations in human cholangiocarcinoma cell lines by cross- species color banding. Genes Chromosomes Cancer. 2001. 30:48–56.21. Scandura JM, Boccuni P, Cammenga J, Nimer SD. Transcription factor fusions in acute leukemia: variations on a theme. Oncogene. 2002. 21:3422–3444.
Article22. Gao FH, Wang Q, Wu YL, Li X, Zhao KW, Chen GQ. c-Jun N-terminal kinase mediates AML1-ETO protein-induced connexin-43 expression. Biochem Biophys Res Commun. 2007. 356:505–511.
Article23. Li X, Xu YB, Wang Q, Lu Y, Zheng Y, Wang YC, Lubbert M, Zhao KW, Chen GQ. Leukemogenic AML1-ETO fusion protein upregulates expression of connexin 43: the role in AML 1-ETO-induced growth arrest in leukemic cells. J Cell Physiol. 2006. 208:594–601.24. Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005. 105:2640–2653.
Article25. Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003. 306:1–12.
Article26. Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood. 2004. 103:4630–4635.
Article27. Chim CS, Wong KY, Loong F, Srivastava G. SOCS1 and SHP1 hypermethylation in mantle cell lymphoma and follicular lymphoma: implications for epigenetic activation of the Jak/STAT pathway. Leukemia. 2004. 18:356–358.
Article28. Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005. 105:2640–2653.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Acute myeloid leukemia arising from chronic myelomonocytic leukemia during hypomethylating therapy
- Granulocytic Sarcoma in the Leg Mimicking Hemorrhagic Abscess
- Acute Myeloid Leukemia with Intracardiac Thrombus Presenting as Acute Limb Ischemia
- A Case of Leukemia Cutis in Myelodysplastic Syndrome Evolving into An Atypical Chronic Myeloid Leukemia
- Myeloid Sarcoma of Peritoneum in Acute Myeloid Leukemia Patient with Inversion of Chromosome 16