J Korean Diabetes Assoc.
2007 May;31(3):193-199. 10.4093/jkda.2007.31.3.193.
Microarray Analysis of Short Heterodimer Partner (SHP)-induced Changes in Gene Expression in INS-1 Cells
- Affiliations
-
- 1Department of Internal Medicine, Catholic University of Daegu School of Medicine, Korea.
- 2Department of Internal Medicine, Kyungpook National University College of Medicine, Korea.
- KMID: 1523007
- DOI: http://doi.org/10.4093/jkda.2007.31.3.193
Abstract
-
BACKGROUND: Nuclear receptors are involved in the cell growth, development, differentiation, and metabolism. The orphan nuclear receptor SHP which lacks a DNA-binding domain is a negative regulator of nuclear receptor signaling pathways. In pancreas, SHP regulate transcriptional activity of HNF3 and HNF4 through binding them and BETA2 which is involved in beta cell differentiation and insulin production. Here, we examined transcriptional activity changes of genes expressed in beta cell when SHP was overexpressed.
METHOD: INS-1 cells of passage number 24 - 30 were prepared. Affimetrix DNA chip was used to examine gene expression in INS-1 cell when SHP was overexpressed. INS-1 cells were infected with adenovirus-SHP to overexpress SHP. To confirm the result of DNA chip, we used real time RT-PCR.
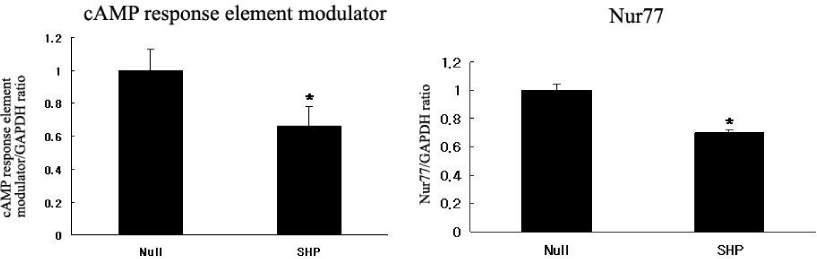
RESULT: When SHP was overexpressed by adenovirus-SHP transfection, FXR, Transforming growth factor, beta 2, fructose-1,6-bisphosphatase 2, bone morphogenetic protein 4 genes expression were increased. Contrarily, Activating transcription factor 2, Glycogen synthase kinase 3 alpha, Nur 77, fibroblast growth factor 14 genes expression were decreased. We confirmed DNA microarray analysis by real time RT-PCR. FXR, tribbles homolog 3 (Drosophila), fructose-1,6-bisphosphatase 2, CD36 genes expression were increased in real time RT-PCR. Nur 77 and cAMP response element modulator genes expression were decreased in real time RT-PCR.
CONCLUSION
we identified several genes which expression are regulated by SHP in pancreas beta cell. These results help to explain how SHP act in the various metabolism of pancreas beta cell.
Keyword
MeSH Terms
-
Activating Transcription Factor 2
Bone Morphogenetic Protein 4
Cell Differentiation
Child
Child, Orphaned
Fibroblast Growth Factors
Fructose-Bisphosphatase
Gene Expression*
Glycogen Synthase Kinase 3
Humans
Insulin
Metabolism
Microarray Analysis*
Oligonucleotide Array Sequence Analysis
Pancreas
Receptors, Cytoplasmic and Nuclear
Response Elements
Transfection
Transforming Growth Factors
Activating Transcription Factor 2
Bone Morphogenetic Protein 4
Fibroblast Growth Factors
Fructose-Bisphosphatase
Glycogen Synthase Kinase 3
Insulin
Receptors, Cytoplasmic and Nuclear
Transforming Growth Factors
Figure
Reference
-
1. O'Malley BW, Buller RE. Herman Beerman lecture. Mechanisms of steroid hormone action. J Invest Dermatol. 1977. 68:1–4.2. Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996. 272:1336–1339.3. Borgius LJ, Steffensen KR, Gustafsson JA, Treuter E. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J Biol Chem. 2002. 277:49761–49766.4. Johansson L, Thomsen JS, Damdimopoulos AE, Spyrou G, Gustafsson JA, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999. 274:345–353.5. Seol W, Hanstein B, Brown M, Moore DD. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner). Mol Endocrinol. 1998. 12:1551–1557.6. Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol. 2000. 20:187–195.7. Sanyal S, Kim JY, Kim HJ, Takeda J, Lee YK, Moore DD, Choi HS. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J Biol Chem. 2002. 277:1739–1748.8. Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol. 2002. 16:2065–2076.9. Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, Vilarem MJ, Pascussi JM. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003. 17:1693–1703.10. Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol. 2005. 19:950–963.11. Jung D, Kullak-Ublick GA. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology. 2003. 37:622–631.12. Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000. 6:517–526.13. Kim HJ, Kim JY, Kim JY, Park SK, Seo JH, Kim JB, Lee IK, Kim KS, Choi HS. Differential regulation of human and mouse orphan nuclear receptor small heterodimer partner promoter by sterol regulatory element binding protein-1. J Biol Chem. 2004. 279:28122–28131.14. Kim JY, Kim HJ, Kim KT, Park YY, Seong HA, Park KC, Lee IK, Ha H, Shong M, Park SC, Choi HS. Orphan nuclear receptor small heterodimer partner represses hepatocyte nuclear factor 3/Foxa transactivation via inhibition of its DNA binding. Mol Endocrinol. 2004. 18:2880–2894.15. Kim JY, Chu K, Kim HJ, Seong HA, Park KC, Sanyal S, Takeda J, Ha H, Shong M, Tsai MJ, Choi HS. Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix-loop-helix transcription factor BETA2/neuroD. Mol Endocrinol. 2004. 18:776–790.16. Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, Kohama T, Yamagata K, Yang Q, Matsuzawa Y, Miki T, Seino S, Kim MY, Choi HS, Lee YK, Moore DD, Takeda J. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci USA. 2001. 98:575–580.17. Mitchell SM, Weedon MN, Owen KR, Shields B, Wilkins-Wall B, Walker M, McCarthy MI, Frayling TM, Hattersley AT. Genetic variation in the small heterodimer partner gene and young-onset type 2 diabetes, obesity, and birth weight in U.K. subjects. Diabetes. 2003. 52:1276–1279.18. He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998. 95:2509–2514.19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods. 2001. 25:402–408.20. Roche E, Buteau J, Aniento I, Reig JA, Soria B, Prentki M. Palmitate and oleate induce the immediate-early response genes c-fos and nur-77 in the pancreatic beta-cell line INS-1. Diabetes. 1999. 48:2007–2014.21. Yew KH, Prasadan KL, Preuett BL, Hembree MJ, McFall CR, Benjes CL, Crowley AR, Sharp SL, Li Z, Tulachan SS, Mehta SS, Gittes GK. Interplay of glucagon-like peptide-1 and transforming growth factor-beta signaling in insulin-positive differentiation of AR42J cells. Diabetes. 2004. 53:2824–2835.22. Yagi N, Yokono K, Amano K, Nagata M, Tsukamoto K, Hasegawa Y, Yoneda R, Okamoto N, Moriyama H, Miki M. Expression of intercellular adhesion molecule 1 on pancreatic beta-cells accelerates beta-cell destruction by cytotoxic T-cells in murine autoimmune diabetes. Diabetes. 1995. 44:744–752.23. Toivonen A, Kulmala P, Rahko J, Ilonen J, Knip M. Soluble adhesion molecules in Finnish schoolchildren with signs of preclinical type 1 diabetes. Diabetes Metab Res Rev. 2004. 20:48–54.24. Lu Y, Herrera PL, Guo Y, Sun D, Tang Z, LeRoith D, Liu JL. Pancreatic-specific inactivation of IGF-I gene causes enlarged pancreatic islets and significant resistance to diabetes. Diabetes. 2004. 53:3131–3141.25. Srinivasan S, Ohsugi M, Liu Z, Fatrai S, Bernal-Mizrachi E, Permutt MA. Endoplasmic reticulum stress-induced apoptosis is partly mediated by reduced insulin signaling through phosphatidy-linositol 3-kinase/Akt and increased glycogen synthase kinase-3beta in mouse insulinoma cells. Diabetes. 2005. 54:968–975.26. Yew KH, Hembree M, Prasadan K, Preuett B, McFall C, Benjes C, Crowley A, Sharp S, Tulachan S, Mehta S, Tei E, Gittes G. Cross-talk between bone morphogenetic protein and transforming growth factor-beta signaling is essential for exendin-4-induced insulin-positive differentiation of AR42J cells. J Biol Chem. 2005. 280:32209–32217.27. Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006. 116:1102–1109.28. Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005. 24:2624–2633.29. Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004. 279:23158–23165.30. Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005. 2:227–238.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Thyroid Hormone Regulates the mRNA Expression of Small Heterodimer Partner through Liver Receptor Homolog-1
- The Analysis of SHP (Small Heterodimer Partner) Gene Mutation in Infertile Patients with Polycystic Ovary Syndrome (PCOS) in Korea
- The orphan nuclear receptor SHP inhibits apoptosis during the monocytic differentiation by inducing p21WAF1
- Roles of Orphan Nuclear Receptor Small Heterodimer Partner in Bone Development: Microcomputed Tomographic Analysis of Bone Microarchitecture in SHP Knockout Mice
- Small Heterodimer Partner and Innate Immune Regulation