J Korean Med Sci.
2025 May;40(19):e69. 10.3346/jkms.2025.40.e69.
Amplicon-Based MinION Sequencing Complements Severe Fever With Thrombocytopenia Syndrome (SFTS) Diagnosis via Real-Time RT-PCR in Patients With Suspected SFTS
- Affiliations
-
- 1Department of Microbiology, Hallym University College of Medicine, Chuncheon, Korea
- 2Center for Antimicrobial Resistance and Microbial Genetics, University of Ulsan College of Medicine, Seoul, Korea
- 3Asan Institute for Life Science, Asan Medical Center, Seoul, Korea
- 4Division of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 5Department of Microbiology, Korea University College of Medicine, Seoul, Korea
- 6BK21 Graduate Program, Biomedical Sciences, Korea University College of Medicine, Seoul, Korea
- 7Division of Infectious Diseases, Department of Internal Medicine, Chuncheon Sacred Heart Hospital, Hallym University College of Medicine, Chuncheon, Korea
- 8Institute of Medical Science, Hallym University College of Medicine, Chuncheon, Korea
- KMID: 2568105
- DOI: http://doi.org/10.3346/jkms.2025.40.e69
Abstract
- Background
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a lethal threat. Increasing Severe fever with thrombocytopenia syndrome (SFTS) risk in Asia and the United States stems from the spread of natural host, Haemaphysalis longicornis. Rapid and accurate SFTSV molecular diagnosis is crucial for treatment decisions, reducing fatality risk.
Methods
Blood samples from 17 suspected SFTS patients at Chuncheon Sacred Heart Hospital (September-December 2022) were collected. SFTSV was diagnosed using two reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays from Gangwon Institute of Health and Environment (RT-qPCR/GIHE) and Asan Medical Center (RT-qPCR/AMC). To address RT-qPCR disparities, amplicon-based MinION sequencing traced SFTSV genomic sequences in clinical samples.
Results
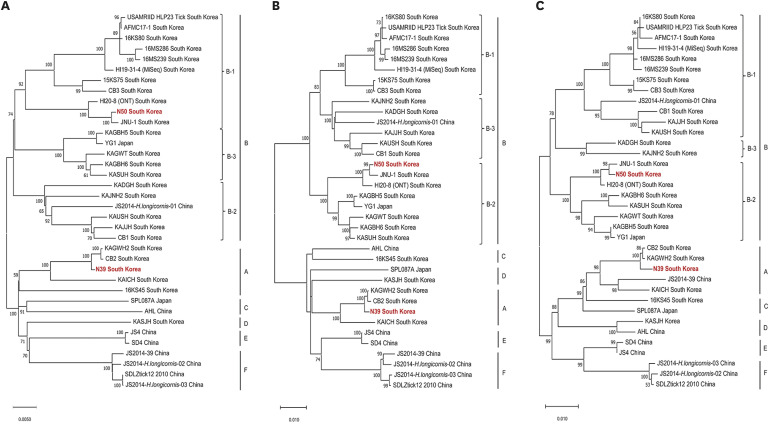
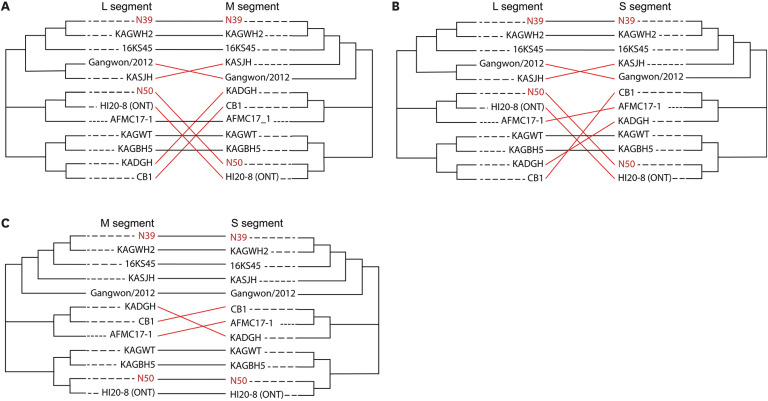
In two samples (N39 and N50), SFTSV was detected in both RT-qPCR/GIHE and RTqPCR/AMC. Among 11 samples, RT-qPCR/AMC exclusively detected SFTSV. In four samples, both assays yielded negative results. Amplicon-based MinION sequencing enabled nearly whole-genome sequencing of SFTSV in samples N39 and N50. Among 11 discordant samples, five contained significant SFTSV reads, aligning with the RT-qPCR/AMC findings. However, another six samples showed insufficient viral reads in accordance with the negativity observed in RT-qPCR/GIHE. The phylogenetic pattern of SFTSV demonstrated N39 formed a genetic lineage with genotype A in all segments. SFTSV N50 grouped with the B-1 sub-genotype for L segment and B-2 sub-genotype for the M and S segments, indicating genetic reassortment.
Conclusion
The study demonstrates the robust sensitivity of amplicon-based MinION sequencing for the direct detection of SFTSV in clinical samples containing ultralow copies of viral genomes. Next-generation sequencing holds potential in resolving SFTSV diagnosis discrepancies, enhancing understanding of diagnostic capacity, and risk assessment for emerging SFTSV.
Keyword
Figure
Reference
-
1. Liu Q, He B, Huang SY, Wei F, Zhu XQ. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect Dis. 2014; 14(8):763–772. PMID: 24837566.2. Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011; 364(16):1523–1532. PMID: 21410387.3. Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, et al. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis. 2013; 19(11):1892–1894. PMID: 24206586.4. Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis. 2014; 209(6):816–827. PMID: 24231186.5. Peng SH, Yang SL, Tang SE, Wang TC, Hsu TC, Su CL, et al. Human case of severe fever with thrombocytopenia syndrome virus infection, Taiwan, 2019. Emerg Infect Dis. 2020; 26(7):1612–1614. PMID: 32568054.6. Hoogstraal H, Roberts FH, Kohls GM, Tipton VJ. Review of Haemaphysalis (Kaiseriana) longicornis Neumann (resurrected) of Australia, New Zealand, New Caledonia, Fiji, Japan, Korea, and Northeastern China and USSR, and its parthenogenetic and bisexual populations (Ixodoidea, Ixodidae). J Parasitol. 1968; 54(6):1197–1213. PMID: 5757695.7. United States Department of Agriculture. National Haemaphysalis longicornis (Asian Longhorned Tick) Situation Report. Washington, D.C., USA: United States Department of Agriculture;2023.8. Raghavan RK, Barker SC, Cobos ME, Barker D, Teo EJM, Foley DH, et al. Potential spatial distribution of the newly introduced long-horned tick, Haemaphysalis longicornis in North America. Sci Rep. 2019; 9(1):498. PMID: 30679711.9. Mehand MS, Al-Shorbaji F, Millett P, Murgue B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. 2018; 159:63–67. PMID: 30261226.10. Seo JW, Kim D, Yun N, Kim DM. Clinical update of severe fever with thrombocytopenia syndrome. Viruses. 2021; 13(7):1213. PMID: 34201811.11. Fu Y, Li S, Zhang Z, Man S, Li X, Zhang W, et al. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan Islands, China: implication for transmission across the ocean. Sci Rep. 2016; 6(1):19563. PMID: 26806841.12. Yun SM, Park SJ, Kim YI, Park SW, Yu MA, Kwon HI, et al. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight. 2020; 5(2):e129531. PMID: 31877113.13. Cui L, Ge Y, Qi X, Xu G, Li H, Zhao K, et al. Detection of severe fever with thrombocytopenia syndrome virus by reverse transcription-cross-priming amplification coupled with vertical flow visualization. J Clin Microbiol. 2012; 50(12):3881–3885. PMID: 22993179.14. Li D. A highly pathogenic new bunyavirus emerged in China. Emerg Microbes Infect. 2013; 2(1):e1. PMID: 26038435.15. Zhang M, Du Y, Yang L, Zhan L, Yang B, Huang X, et al. Development of monoclonal antibody based IgG and IgM ELISA for diagnosis of severe fever with thrombocytopenia syndrome virus infection. Braz J Infect Dis. 2022; 26(4):102386. PMID: 35835158.16. Zhan L, Huang K, Xia W, Chen J, Wang L, Lu J, et al. The diagnosis of severe fever with thrombocytopenia syndrome using metagenomic next-generation sequencing: case report and literature review. Infect Drug Resist. 2022; 15:83–89. PMID: 35046673.17. Baek YH, Cheon HS, Park SJ, Lloren KKS, Ahn SJ, Jeong JH, et al. Simple, rapid and sensitive portable molecular diagnosis of SFTS virus using reverse transcriptional loop-mediated isothermal amplification (RT-LAMP). J Microbiol Biotechnol. 2018; 28(11):1928–1936. PMID: 30270605.18. Yoshikawa T, Fukushi S, Tani H, Fukuma A, Taniguchi S, Toda S, et al. Sensitive and specific PCR systems for detection of both Chinese and Japanese severe fever with thrombocytopenia syndrome virus strains and prediction of patient survival based on viral load. J Clin Microbiol. 2014; 52(9):3325–3333. PMID: 24989600.19. Park ES, Fujita O, Kimura M, Hotta A, Imaoka K, Shimojima M, et al. Diagnostic system for the detection of severe fever with thrombocytopenia syndrome virus RNA from suspected infected animals. PLoS One. 2021; 16(1):e0238671. PMID: 33507990.20. Li Z, Qi X, Zhou M, Bao C, Hu J, Wu B, et al. A two-tube multiplex real-time RT-PCR assay for the detection of four hemorrhagic fever viruses: severe fever with thrombocytopenia syndrome virus, Hantaan virus, Seoul virus, and dengue virus. Arch Virol. 2013; 158(9):1857–1863. PMID: 23532380.21. Sun Y, Liang M, Qu J, Jin C, Zhang Q, Li J, et al. Early diagnosis of novel SFTS bunyavirus infection by quantitative real-time RT-PCR assay. J Clin Virol. 2012; 53(1):48–53. PMID: 22024488.22. Jalal S, Hwang SY, Kim CM, Kim DM, Yun NR, Seo JW, et al. Comparison of RT-PCR, RT-nested PCRs, and real-time PCR for diagnosis of severe fever with thrombocytopenia syndrome: a prospective study. Sci Rep. 2021; 11(1):16764. PMID: 34408188.23. Kwon JS, Kim MC, Kim JY, Jeon NY, Ryu BH, Hong J, et al. Kinetics of viral load and cytokines in severe fever with thrombocytopenia syndrome. J Clin Virol. 2018; 101:57–62. PMID: 29427908.24. Huang X, Li J, Li A, Wang S, Li D. Epidemiological characteristics of severe fever with thrombocytopenia syndrome from 2010 to 2019 in Mainland China. Int J Environ Res Public Health. 2021; 18(6):3092. PMID: 33802869.25. Kim JY, Koo B, Jin CE, Kim MC, Chong YP, Lee SO, et al. Rapid diagnosis of tick-borne illnesses by use of one-step isothermal nucleic acid amplification and bio-optical sensor detection. Clin Chem. 2018; 64(3):556–565. PMID: 29208659.26. Lee J, Park K, Kim J, Lee SH, Lee GY, Cho S, et al. Whole-genome sequencing and genetic diversity of severe fever with thrombocytopenia syndrome virus using multiplex PCR-based nanopore sequencing, Republic of Korea. PLoS Negl Trop Dis. 2022; 16(9):e0010763. PMID: 36094957.27. Nagarajan N, Kingsford C. GiRaF: robust, computational identification of influenza reassortments via graph mining. Nucleic Acids Res. 2011; 39(6):e34. PMID: 21177643.28. Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015; 31(22):3718–3720. PMID: 26209431.29. Sharma D, Kamthania M. A new emerging pandemic of severe fever with thrombocytopenia syndrome (SFTS). Virusdisease. 2021; 32(2):220–227. PMID: 33942022.30. Wick RR, Judd LM, Holt KE. Deepbinner: demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLOS Comput Biol. 2018; 14(11):e1006583. PMID: 30458005.31. Brandt C, Krautwurst S, Spott R, Lohde M, Jundzill M, Marquet M, et al. poreCov-an easy to use, fast, and robust workflow for SARS-CoV-2 genome reconstruction via nanopore sequencing. Front Genet. 2021; 12:711437. PMID: 34394197.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison Between the SFTS-QS Kit and the PowerChek SFTSV Real-time PCR Kit for the Detection of Severe Fever With Thrombocytopenia Syndrome Virus
- Four Cases of Severe Fever with Thrombocytopenia Syndrome Occurring in Jeju
- Current Status and Infection Control of Severe Fever with Thrombocytopenia Syndrome in Korea
- The First Case of Non-retrospective Clinical Identification of Severe Fever with Thrombocytopenia Syndrome Patient in 2013 in South Korea
- Severe Fever with Thrombocytopenia Syndrome Accompanied by Hemophagocytic Lymphohistiocytosis