Brain Tumor Res Treat.
2024 Oct;12(4):237-244. 10.14791/btrt.2024.0035.
The Inflammatory Characteristics of Symptomatic Glioma Associated With Poor Prognosis and Chemoresistance via Tumor Necrosis Factor Signaling Pathway
- Affiliations
-
- 1Department of Biomedical Science, College of Life Science, CHA University, Seongnam, Korea
- 2Department of Medicine, College of Medicine, Hallym University, Chuncheon, Korea
- 3CHA Institute for Future Medicine, Medical Center Research Institute, Seongnam, Korea
- 4Department of Neurosurgery, Bundang CHA Medical Center, CHA University College of Medicine, Seongnam, Korea
- 5Department of Neurosurgery, Brain Tumor Center, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 6Department of Neurosurgery, Dong-A University Hospital, Dong-A University College of Medicine, Busan, Korea
- 7Department of Neurosurgery, Brain Tumor Center, Gangnam Severance Hospital, Seoul, Korea
- KMID: 2561288
- DOI: http://doi.org/10.14791/btrt.2024.0035
Abstract
- Background
Among gliomas, the most common primary malignant brain tumor, incidental gliomas account for 2.5%–5% of cases. The controversy over whether to pursue immediate treatment or adopt a wait-and-see approach remains, and more molecular and immunological evidence is needed for definitive treatment decisions.
Methods
Total RNA sequencing (RNA-seq) data and single cell RNA sequencing (scRNA-seq) data were retrospectively analyzed to compare the molecular and immunological tumor microenvironment differences between incidental glioma and symptomatic glioma samples. These were classified using symptom data from The Cancer Genome Atlas (TCGA) and public dataset.
Results
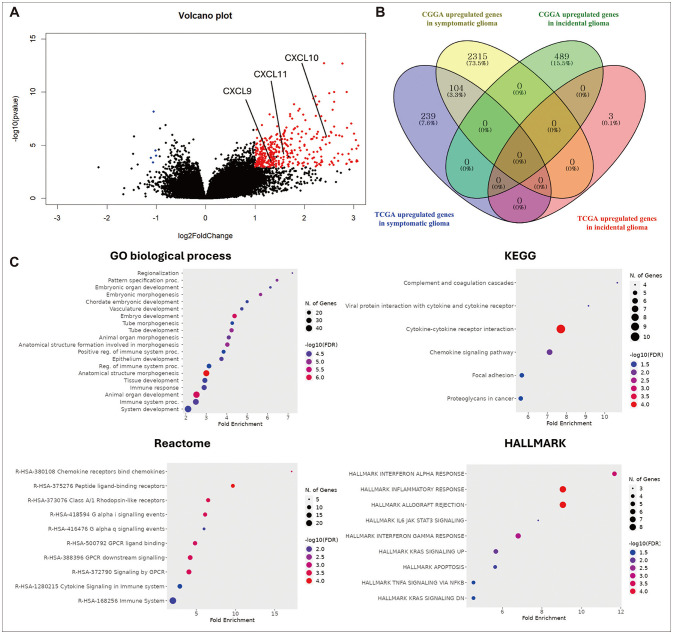
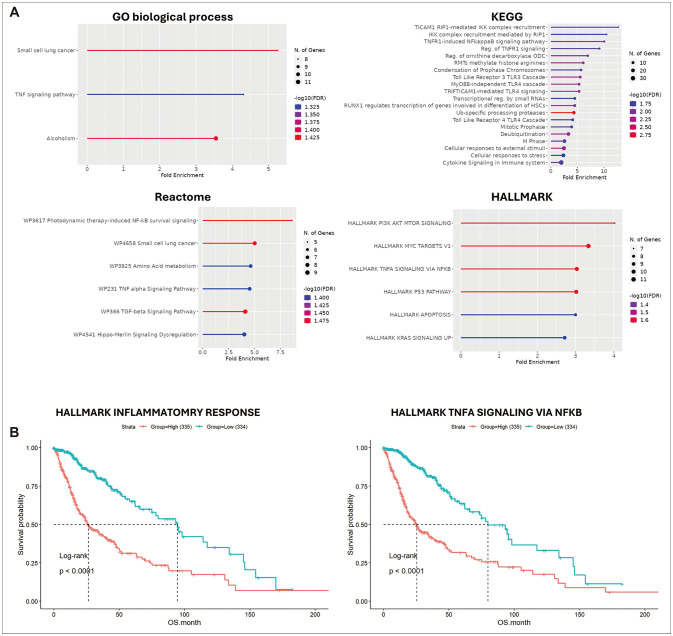
RNA-seq analysis of the GBMLGG dataset identified 343 genes upregulated in symp- tomatic glioma and 118 in incidental glioma, with 104 common genes upregulated in symptomatic glioma across both the TCGA and Chinese Glioma Genome Atlas (CGGA) datasets. Enrichment analysis revealed that these 104 genes in symptomatic glioma were significantly associated with immunological pathways. scRNA-seq analysis of glioma revealed 11 cell types, including T cells, myeloid cells, and oligodendrocytes, with the tumor necrosis factor (TNF) signaling pathway strongly influencing other cell types, particularly myeloid cells. Enrichment and survival analyses showed that TNF signaling is associated with temozolomide resistance and poorer prognosis in glioma patients.
Conclusion
The findings suggest that symptomatic glioma enhances inflammatory responses linked to poor prognosis and chemoresistance. This supports the hypothesis that immediate treatment of incidental glioma may improve patient outcomes over a wait-and-see approach.
Keyword
Figure
Reference
-
1. Ho VK, Reijneveld JC, Enting RH, Bienfait HP, Robe P, Baumert BG, et al. Changing incidence and improved survival of gliomas. Eur J Cancer. 2014; 50:2309–2318. PMID: 24972545.
Article2. Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS. Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018; 4:1254–1262. PMID: 29931168.
Article3. Posti JP, Bori M, Kauko T, Sankinen M, Nordberg J, Rahi M, et al. Presenting symptoms of glioma in adults. Acta Neurol Scand. 2015; 131:88–93. PMID: 25263022.
Article4. Wen PY, Reardon DA. Neuro-oncology in 2015: progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016; 12:69–70. PMID: 26782337.
Article5. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021; 23:1231–1251. PMID: 34185076.
Article6. Taal W, Bromberg JE, van den Bent MJ. Chemotherapy in glioma. CNS Oncol. 2015; 4:179–192. PMID: 25906059.
Article7. Pallud J, Fontaine D, Duffau H, Mandonnet E, Sanai N, Taillandier L, et al. Natural history of incidental World Health Organization grade II gliomas. Ann Neurol. 2010; 68:727–733. PMID: 21031584.
Article8. Ius T, Cesselli D, Isola M, Pauletto G, Tomasino B, D’Auria S, et al. Incidental low-grade gliomas: single-institution management based on clinical, surgical, and molecular data. Neurosurgery. 2020; 86:391–399. PMID: 31260076.
Article9. Zeng L, Mei Q, Li H, Ke C, Yu J, Chen J. A survival analysis of surgically treated incidental low-grade glioma patients. Sci Rep. 2021; 11:8522. PMID: 33875775.
Article10. Hayhurst C, Mendelsohn D, Bernstein M. Low grade glioma: a qualitative study of the wait and see approach. Can J Neurol Sci. 2011; 38:256–261. PMID: 21320830.
Article11. Allison CM, Shumon S, Stummer W, Holling M, Surash S. A cohort analysis of truly incidental low-grade gliomas. World Neurosurg. 2022; 159:e347–e355. PMID: 34942387.
Article12. Potts MB, Smith JS, Molinaro AM, Berger MS. Natural history and surgical management of incidentally discovered low-grade gliomas. J Neurosurg. 2012; 116:365–372. PMID: 21999317.
Article13. Zhang ZY, Chan AK, Ng HK, Ding XJ, Li YX, Shi ZF, et al. Surgically treated incidentally discovered low-grade gliomas are mostly IDH mutated and 1p19q co-deleted with favorable prognosis. Int J Clin Exp Pathol. 2014; 7:8627–8636. PMID: 25674227.14. Park J, Sim J, Ahn J, Kim YJ, Hwang S, Cho K, et al. Molecular characteristics of incidental lower-grade glioma for treatment decision-making. J Neurosurg. 2022; 138:629–638. PMID: 35986732.
Article15. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. PMID: 25516281.
Article16. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 2024; 42:293–304. PMID: 37231261.
Article17. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021; 12:1088. PMID: 33597522.
Article18. Wang QW, Wang YW, Wang ZL, Bao ZS, Jiang T, Wang Z, et al. Clinical and molecular characterization of incidentally discovered lower-grade gliomas with enrichment of aerobic respiration. Onco Targets Ther. 2020; 13:9533–9542. PMID: 33061437.19. Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019; 10:317. PMID: 31024627.
Article20. Kharchenko PV. The triumphs and limitations of computational methods for scRNA-seq. Nat Methods. 2021; 18:723–732. PMID: 34155396.
Article21. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017; 97:498–518. PMID: 28287634.
Article22. Ghouzlani A, Kandoussi S, Tall M, Reddy KP, Rafii S, Badou A. Immune checkpoint inhibitors in human glioma microenvironment. Front Immunol. 2021; 12:679425. PMID: 34305910.
Article23. Lin W, Wu S, Chen X, Ye Y, Weng Y, Pan Y, et al. Characterization of hypoxia signature to evaluate the tumor immune microenvironment and predict prognosis in glioma groups. Front Oncol. 2020; 10:796. PMID: 32500034.
Article24. Sim J, Ahn JW, Park J, Kim YJ, Jeong JY, Lee JM, et al. Non-canonical NLRC4 inflammasomes in astrocytes contribute to glioma malignancy. Inflamm Res. 2023; 72:813–827. PMID: 36899084.
Article25. Sim J, Park J, Kim S, Hwang S, Sung K, Lee JE, et al. Association of Tim-3/Gal-9 axis with NLRC4 inflammasome in glioma malignancy: Tim-3/Gal-9 induce the NLRC4 inflammasome. Int J Mol Sci. 2022; 23:2028. PMID: 35216164.
Article26. Park J, Kim YJ, Lee M, Kim D, Sim J, Cho K, et al. Correlation of LLT-1 and NLRC4 inflammasome and its effect on glioblastoma prognosis. J Neurooncol. 2024; 169:543–553. PMID: 38907949.
Article27. Zhang H, Luo YB, Wu W, Zhang L, Wang Z, Dai Z, et al. The molecular feature of macrophages in tumor immune microenvironment of glioma patients. Comput Struct Biotechnol J. 2021; 19:4603–4618. PMID: 34471502.
Article28. Price G, Bouras A, Hambardzumyan D, Hadjipanayis CG. Current knowledge on the immune microenvironment and emerging immunotherapies in diffuse midline glioma. EBioMedicine. 2021; 69:103453. PMID: 34157482.
Article29. Ha ET, Antonios JP, Soto H, Prins RM, Yang I, Kasahara N, et al. Chronic inflammation drives glioma growth: cellular and molecular factors responsible for an immunosuppressive microenvironment. Neuroimmunol Neuroinflammation. 2014; 1:66–76.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- IGF-I Exerts an Anti-inflammatory Effect on Skeletal Muscle Cells through Down-regulation of TLR4 Signaling
- The Role of the Ubiquitin-Proteasome Pathway in Neurodegenerative Disorders
- Impact of Nrf2 overexpression on cholangiocarcinoma treatment and clinical prognosis
- Role of Reactive Oxygen Species in Cell Death Pathways
- The Dose Dependent Effects of Ruxolitinib on the Invasion and Tumorigenesis in Gliomas Cells via Inhibition of Interferon Gamma-Depended JAK/STAT Signaling Pathway