Diabetes Metab J.
2024 May;48(3):340-353. 10.4093/dmj.2023.0174.
Roles of Histone Deacetylase 4 in the Inflammatory and Metabolic Processes
- Affiliations
-
- 1Department of Food and Nutrition, Keimyung University, Daegu, Korea
- 2Department of Nutritional Sciences, University of Connecticut, Storrs, CT, USA
- 3Department of Food and Nutrition, Yonsei University, Seoul, Korea
- KMID: 2555766
- DOI: http://doi.org/10.4093/dmj.2023.0174
Abstract
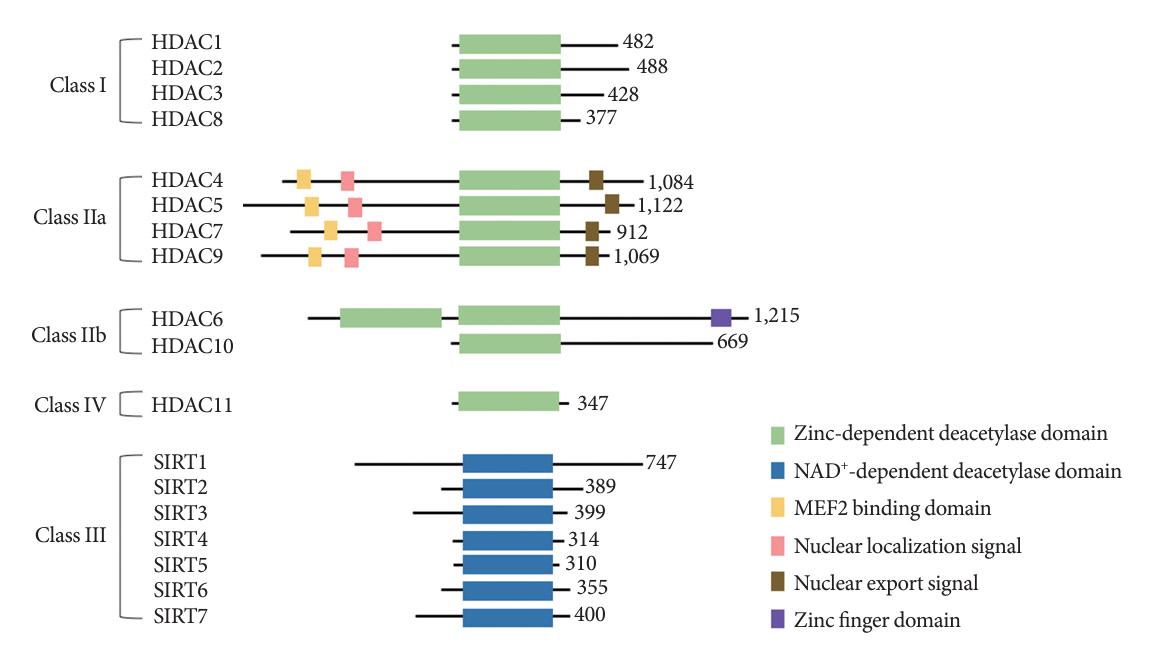
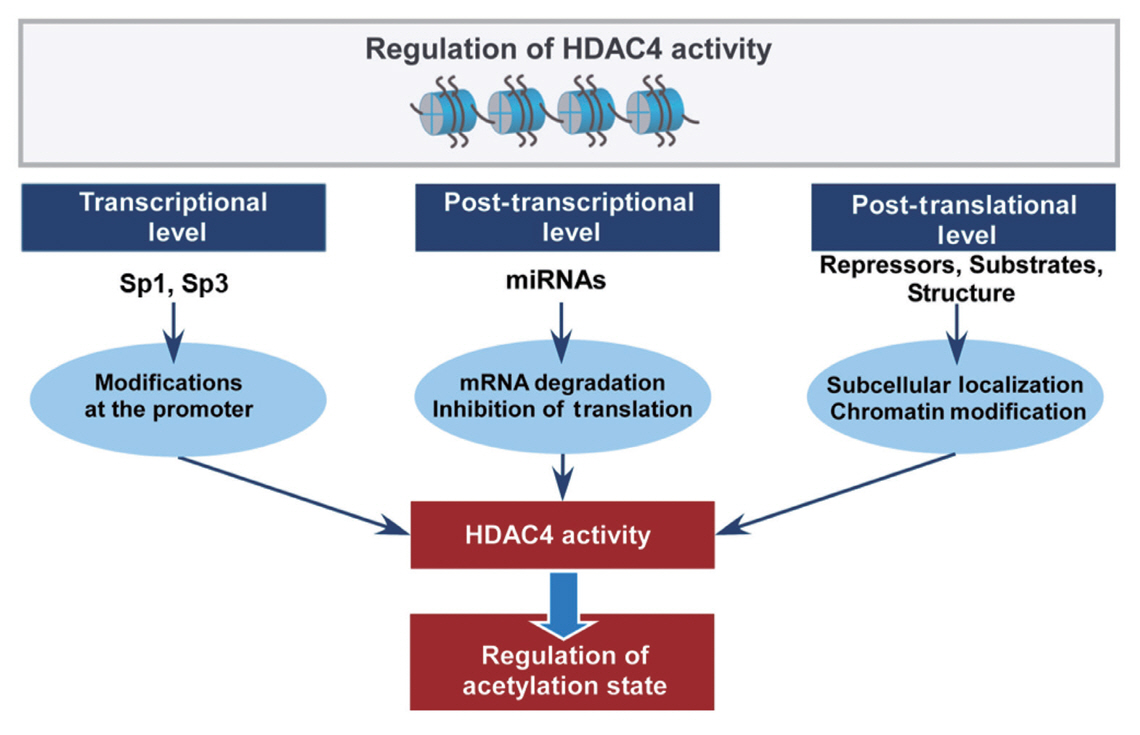
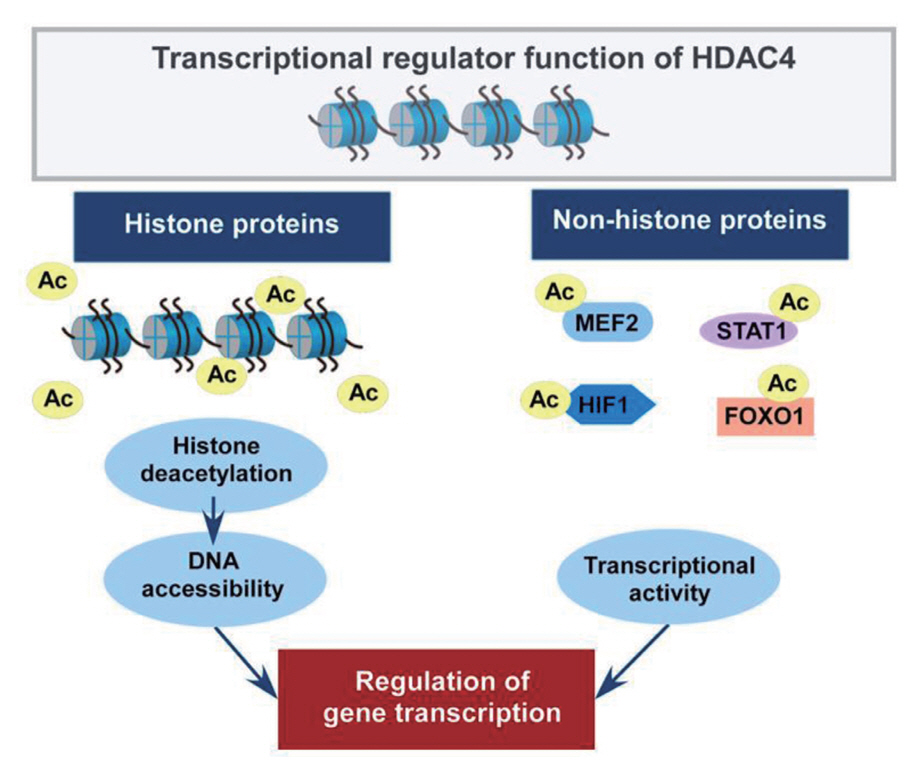
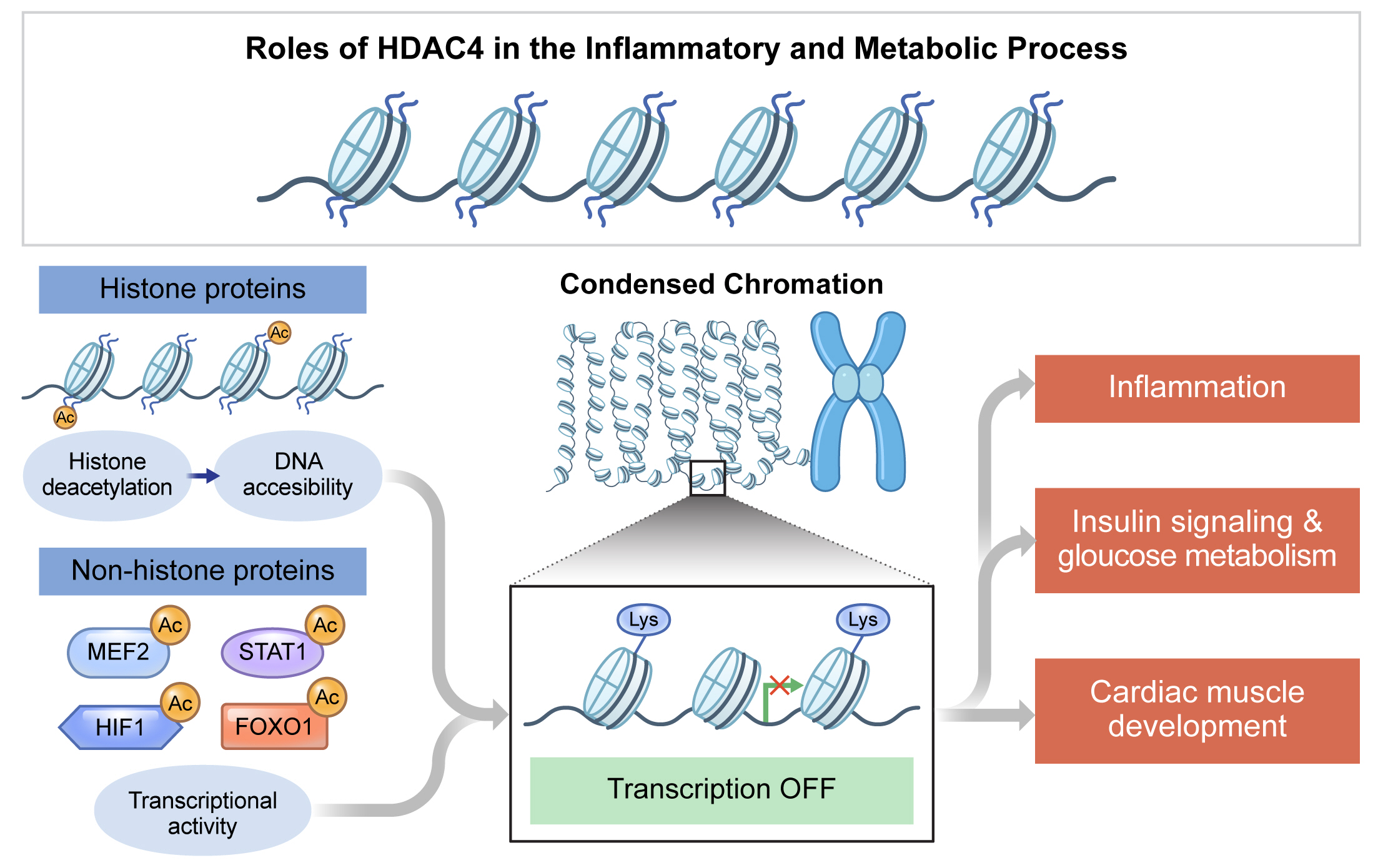
- Histone deacetylase 4 (HDAC4), a class IIa HDAC, has gained attention as a potential therapeutic target in treating inflammatory and metabolic processes based on its essential role in various biological pathways by deacetylating non-histone proteins, including transcription factors. The activity of HDAC4 is regulated at the transcriptional, post-transcriptional, and post-translational levels. The functions of HDAC4 are tissue-dependent in response to endogenous and exogenous factors and their substrates. In particular, the association of HDAC4 with non-histone targets, including transcription factors, such as myocyte enhancer factor 2, hypoxia-inducible factor, signal transducer and activator of transcription 1, and forkhead box proteins, play a crucial role in regulating inflammatory and metabolic processes. This review summarizes the regulatory modes of HDAC4 activity and its functions in inflammation, insulin signaling and glucose metabolism, and cardiac muscle development.
Figure
Reference
-
1. Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014; 6:139–50.
Article2. Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293:1074–80.
Article3. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–95.
Article4. Mielcarek M, Zielonka D, Carnemolla A, Marcinkowski JT, Guidez F. HDAC4 as a potential therapeutic target in neurodegenerative diseases: a summary of recent achievements. Front Cell Neurosci. 2015; 9:42.
Article5. Makinistoglu MP, Karsenty G. The class II histone deacetylase HDAC4 regulates cognitive, metabolic and endocrine functions through its expression in osteoblasts. Mol Metab. 2014; 4:64–9.
Article6. Peng L, Seto E. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb Exp Pharmacol. 2011; 206:39–56.
Article7. Clocchiatti A, Di Giorgio E, Demarchi F, Brancolini C. Beside the MEF2 axis: unconventional functions of HDAC4. Cell Signal. 2013; 25:269–76.
Article8. Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol. 2001; 79:243–52.
Article9. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014; 6:a018713.
Article10. Chen H, Xie C, Chen Q, Zhuang S. HDAC11, an emerging therapeutic target for metabolic disorders. Front Endocrinol (Lausanne). 2022; 13:989305.
Article11. Makkar R, Behl T, Arora S. Role of HDAC inhibitors in diabetes mellitus. Curr Res Transl Med. 2020; 68:45–50.
Article12. Bagchi RA, Weeks KL. Histone deacetylases in cardiovascular and metabolic diseases. J Mol Cell Cardiol. 2019; 130:151–9.
Article13. Shukla S, Tekwani BL. Histone deacetylases inhibitors in neurodegenerative diseases, neuroprotection and neuronal differentiation. Front Pharmacol. 2020; 11:537.
Article14. Pham TX, Park YK, Lee JY. Anti-inflammatory effects of spirulina platensis extract via the modulation of histone deacetylases. Nutrients. 2016; 8:381.
Article15. Luan B, Goodarzi MO, Phillips NG, Guo X, Chen YD, Yao J, et al. Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab. 2014; 19:1058–65.
Article16. Du J, Zhang L, Wang H, Zhao YT, Yano N, Zhao TC. Cardiac specific overexpression of histone deacetylase 4 aggravates high fat diet-induced cardiac dysfunction and metabolic disorders. Circulation. 2016; 134(Suppl 1):A17970.17. Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009; 323:256–9.
Article18. Blixt NC, Faulkner BK, Astleford K, Lelich R, Schering J, Spencer E, et al. Class II and IV HDACs function as inhibitors of osteoclast differentiation. PLoS One. 2017; 12:e0185441.
Article19. Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, et al. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med. 2012; 18:783–90.
Article20. Luo L, Martin SC, Parkington J, Cadena SM, Zhu J, Ibebunjo C, et al. HDAC4 controls muscle homeostasis through deacetylation of myosin heavy chain, PGC-1α, and Hsc70. Cell Rep. 2019; 29:749–63.
Article21. Fitzsimons HL. The class IIa histone deacetylase HDAC4 and neuronal function: nuclear nuisance and cytoplasmic stalwart? Neurobiol Learn Mem. 2015; 123:149–58.
Article22. Borghi S, Molinari S, Razzini G, Parise F, Battini R, Ferrari S. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J Cell Sci. 2001; 114(Pt 24):4477–83.
Article23. Guo L, Han A, Bates DL, Cao J, Chen L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proc Natl Acad Sci U S A. 2007; 104:4297–302.
Article24. Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008; 283:26694–704.
Article25. Porter NJ, Christianson DW. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr Opin Struct Biol. 2019; 59:9–18.
Article26. Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011; 145:607–21.
Article27. Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001; 21:6091–101.
Article28. Lee HA, Song MJ, Seok YM, Kang SH, Kim SY, Kim I. Histone deacetylase 3 and 4 complex stimulates the transcriptional activity of the mineralocorticoid receptor. PLoS One. 2015; 10:e0136801.
Article29. Ginnan R, Sun LY, Schwarz JJ, Singer HA. MEF2 is regulated by CaMKIIδ2 and a HDAC4-HDAC5 heterodimer in vascular smooth muscle cells. Biochem J. 2012; 444:105–14.
Article30. Liu F, Pore N, Kim M, Voong KR, Dowling M, Maity A, et al. Regulation of histone deacetylase 4 expression by the SP family of transcription factors. Mol Biol Cell. 2006; 17:585–97.
Article31. Kang H, Park YK, Lee JY. Inhibition of alcohol-induced inflammation and oxidative stress by astaxanthin is mediated by its opposite actions in the regulation of sirtuin 1 and histone deacetylase 4 in macrophages. Biochim Biophys Acta Mol Cell Biol Lipids. 2021; 1866:158838.
Article32. Okazaki M, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, Nakayama S, et al. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocr J. 2010; 57:403–13.
Article33. Huang ZP, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res. 2013; 112:1234–43.
Article34. Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. MicroRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010; 103:1215–20.
Article35. Lu Y, Li Z, Xie B, Song Y, Ye X, Liu P. hsa-miR-20a-5p attenuates allergic inflammation in HMC-1 cells by targeting HDAC4. Mol Immunol. 2019; 107:84–90.
Article36. Song J, Jin EH, Kim D, Kim KY, Chun CH, Jin EJ. MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during osteoarthritis pathogenesis. BBA Clin. 2014; 3:79–89.
Article37. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011; 54:2025–35.
Article38. Shao L, Hou C. miR-138 activates NF-κB signaling and PGRN to promote rheumatoid arthritis via regulating HDAC4. Biochem Biophys Res Commun. 2019; 519:166–71.
Article39. Chen R, Qiu H, Tong Y, Liao F, Hu X, Qiu Y, et al. MiRNA19a-3p alleviates the progression of osteoporosis by targeting HDAC4 to promote the osteogenic differentiation of hMSCs. Biochem Biophys Res Commun. 2019; 516:666–72.
Article40. Mielcarek M, Landles C, Weiss A, Bradaia A, Seredenina T, Inuabasi L, et al. HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 2013; 11:e1001717.
Article41. Nishino TG, Miyazaki M, Hoshino H, Miwa Y, Horinouchi S, Yoshida M. 14-3-3 regulates the nuclear import of class IIa histone deacetylases. Biochem Biophys Res Commun. 2008; 377:852–6.
Article42. Wu Q, Yang X, Zhang L, Zhang Y, Feng L. Nuclear accumulation of histone deacetylase 4 (HDAC4) exerts neurotoxicity in models of Parkinson’s disease. Mol Neurobiol. 2017; 54:6970–83.
Article43. Litke C, Bading H, Mauceri D. Histone deacetylase 4 shapes neuronal morphology via a mechanism involving regulation of expression of vascular endothelial growth factor D. J Biol Chem. 2018; 293:8196–207.
Article44. Paroni G, Cernotta N, Dello Russo C, Gallinari P, Pallaoro M, Foti C, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008; 19:655–67.
Article45. Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006; 116:1853–64.
Article46. Kreusser MM, Backs J. Integrated mechanisms of CaMKII-dependent ventricular remodeling. Front Pharmacol. 2014; 5:36.
Article47. Liu Y, Schneider MF. Opposing HDAC4 nuclear fluxes due to phosphorylation by β-adrenergic activated protein kinase A or by activity or Epac activated CaMKII in skeletal muscle fibres. J Physiol. 2013; 591:3605–23.
Article48. Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, et al. Selective repression of MEF2 activity by PKAdependent proteolysis of HDAC4. J Cell Biol. 2011; 195:403–15.
Article49. Cernotta N, Clocchiatti A, Florean C, Brancolini C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell. 2011; 22:278–89.
Article50. Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001; 276:35368–74.
Article51. Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002; 21:2682–91.
Article52. Kong Q, Hao Y, Li X, Wang X, Ji B, Wu Y. HDAC4 in ischemic stroke: mechanisms and therapeutic potential. Clin Epigenetics. 2018; 10:117.
Article53. Zhang P, Sun Q, Zhao C, Ling S, Li Q, Chang YZ, et al. HDAC4 protects cells from ER stress induced apoptosis through interaction with ATF4. Cell Signal. 2014; 26:556–63.
Article54. Sando R 3rd, Gounko N, Pieraut S, Liao L, Yates J 3rd, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012; 151:821–34.
Article55. Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013; 14:347–59.
Article56. Liu J, Zhou X, Li Q, Zhou SM, Hu B, Hu GW, et al. Role of phosphorylated HDAC4 in stroke-induced angiogenesis. Biomed Res Int. 2017; 2017:2957538.
Article57. Li L, Yang XJ. Molecular and functional characterization of histone deacetylase 4 (HDAC4). Methods Mol Biol. 2016; 1436:31–45.
Article58. Tao H, Shi KH, Yang JJ, Huang C, Zhan HY, Li J. Histone deacetylases in cardiac fibrosis: current perspectives for therapy. Cell Signal. 2014; 26:521–7.
Article59. Paroni G, Fontanini A, Cernotta N, Foti C, Gupta MP, Yang XJ, et al. Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol Cell Biol. 2007; 27:6718–32.
Article60. Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005; 25:2273–87.
Article61. Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001; 29:3439–47.
Article62. Bolger TA, Zhao X, Cohen TJ, Tsai CC, Yao TP. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem. 2007; 282:29186–92.
Article63. Ciccarelli M, Vastolo V, Albano L, Lecce M, Cabaro S, Liotti A, et al. Glucose-induced expression of the homeotic transcription factor Prep1 is associated with histone post-translational modifications in skeletal muscle. Diabetologia. 2016; 59:176–86.
Article64. Paroni G, Mizzau M, Henderson C, Del Sal G, Schneider C, Brancolini C. Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol Biol Cell. 2004; 15:2804–18.
Article65. Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005; 25:8456–64.
Article66. Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006; 281:4423–33.
Article67. Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003; 3:721–32.
Article68. Shi YH, Fang WG. Hypoxia-inducible factor-1 in tumour angiogenesis. World J Gastroenterol. 2004; 10:1082–7.
Article69. Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001; 15:2675–86.70. Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009; 583:55–60.
Article71. Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010; 38:864–78.72. Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006; 66:8814–21.73. Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, et al. HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011; 286:38095–102.
Article74. Chen S, Sang N. Hypoxia-inducible factor-1: a critical player in the survival strategy of stressed cells. J Cell Biochem. 2016; 117:267–78.
Article75. Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009; 23:223–35.
Article76. Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006; 20:473–85.77. Kaowinn S, Kaewpiboon C, Koh SS, Kramer OH, Chung YH. STAT1-HDAC4 signaling induces epithelial-mesenchymal transition and sphere formation of cancer cells overexpressing the oncogene, CUG2. Oncol Rep. 2018; 40:2619–27.
Article78. Kaewpiboon C, Srisuttee R, Malilas W, Moon J, Oh S, Jeong HG, et al. Upregulation of Stat1-HDAC4 confers resistance to etoposide through enhanced multidrug resistance 1 expression in human A549 lung cancer cells. Mol Med Rep. 2015; 11:2315–21.
Article79. Stronach EA, Alfraidi A, Rama N, Datler C, Studd JB, Agarwal R, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 2011; 71:4412–22.
Article80. Park EJ, Kim YM, Kim HJ, Chang KC. Degradation of histone deacetylase 4 via the TLR4/JAK/STAT1 signaling pathway promotes the acetylation of high mobility group box 1 (HMGB1) in lipopolysaccharide-activated macrophages. FEBS Open Bio. 2018; 8:1119–26.
Article81. Lee JH, Park SM, Kim OS, Lee CS, Woo JH, Park SJ, et al. Differential SUMOylation of LXRalpha and LXRbeta mediates transrepression of STAT1 inflammatory signaling in IFN-gamma-stimulated brain astrocytes. Mol Cell. 2009; 35:806–17.82. Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012; 1819:707–15.
Article83. Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008; 16:1407–16.
Article84. Mosley AL, Ozcan S. Glucose regulates insulin gene transcription by hyperacetylation of histone h4. J Biol Chem. 2003; 278:19660–6.
Article85. Cho HM, Seok YM, Lee HA, Song M, Kim I. Repression of transcriptional activity of forkhead box O1 by histone deacetylase inhibitors ameliorates hyperglycemia in type 2 diabetic rats. Int J Mol Sci. 2018; 19:3539.
Article86. Zhao H, Shu L, Huang W, Song G, Ma H. Resveratrol affects hepatic gluconeogenesis via histone deacetylase 4. Diabetes Metab Syndr Obes. 2019; 12:401–11.87. Gong M, Yu Y, Liang L, Vuralli D, Froehler S, Kuehnen P, et al. HDAC4 mutations cause diabetes and induce β-cell FoxO1 nuclear exclusion. Mol Genet Genomic Med. 2019; 7:e602.88. Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, et al. A hormone-dependent module regulating energy balance. Cell. 2011; 145:596–606.
Article89. Wang B, Liu TY, Lai CH, Rao YH, Choi MC, Chi JT, et al. Glycolysis-dependent histone deacetylase 4 degradation regulates inflammatory cytokine production. Mol Biol Cell. 2014; 25:3300–7.
Article90. Usui T, Okada M, Mizuno W, Oda M, Ide N, Morita T, et al. HDAC4 mediates development of hypertension via vascular inflammation in spontaneous hypertensive rats. Am J Physiol Heart Circ Physiol. 2012; 302:H1894–904.
Article91. Guo Y, Fang Q, Ma D, Yu K, Cheng B, Tang S, et al. Up-regulation of HO-1 promotes resistance of B-cell acute lymphocytic leukemia cells to HDAC4/5 inhibitor LMK-235 via the Smad7 pathway. Life Sci. 2018; 207:386–94.
Article92. Choi MC, Cohen TJ, Barrientos T, Wang B, Li M, Simmons BJ, et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012; 47:122–32.
Article93. Yang Q, Tang J, Pei R, Gao X, Guo J, Xu C, et al. Host HDAC4 regulates the antiviral response by inhibiting the phosphorylation of IRF3. J Mol Cell Biol. 2019; 11:158–69.
Article94. Shakespear MR, Halili MA, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011; 32:335–43.
Article95. O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors: redefining innate immunity. Nat Rev Immunol. 2013; 13:453–60.96. Rossol M, Heine H, Meusch U, Quandt D, Klein C, Sweet MJ, et al. LPS-induced cytokine production in human monocytes and macrophages. Crit Rev Immunol. 2011; 31:379–446.
Article97. Abu-Farha M, Tiss A, Abubaker J, Khadir A, Al-Ghimlas F, Al-Khairi I, et al. Proteomics analysis of human obesity reveals the epigenetic factor HDAC4 as a potential target for obesity. PLoS One. 2013; 8:e75342.
Article98. Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005; 11:191–8.
Article99. Yang Q, Tang J, Xu C, Zhao H, Zhou Y, Wang Y, et al. Histone deacetylase 4 inhibits NF-κB activation by facilitating IκBα sumoylation. J Mol Cell Biol. 2020; 12:933–45.
Article100. Denys A, Udalova IA, Smith C, Williams LM, Ciesielski CJ, Campbell J, et al. Evidence for a dual mechanism for IL-10 suppression of TNF-alpha production that does not involve inhibition of p38 mitogen-activated protein kinase or NF-kappa B in primary human macrophages. J Immunol. 2002; 168:4837–45.101. Ozcan L, Ghorpade DS, Zheng Z, de Souza JC, Chen K, Bessler M, et al. Hepatocyte DACH1 is increased in obesity via nuclear exclusion of HDAC4 and promotes hepatic insulin resistance. Cell Rep. 2016; 15:2214–25.
Article102. Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007; 130:456–69.
Article103. Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R, et al. Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011; 60:2861–71.
Article104. Henriksson E, Sall J, Gormand A, Wasserstrom S, Morrice NA, Fritzen AM, et al. SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. J Cell Sci. 2015; 128:472–86.105. Weems JC, Griesel BA, Olson AL. Class II histone deacetylases downregulate GLUT4 transcription in response to increased cAMP signaling in cultured adipocytes and fasting mice. Diabetes. 2012; 61:1404–14.106. Niu Y, Wang T, Liu S, Yuan H, Li H, Fu L. Exercise-induced GLUT4 transcription via inactivation of HDAC4/5 in mouse skeletal muscle in an AMPKα2-dependent manner. Biochim Biophys Acta Mol Basis Dis. 2017; 1863:2372–81.
Article107. Karamboulas C, Swedani A, Ward C, Al-Madhoun AS, Wilton S, Boisvenue S, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci. 2006; 119(Pt 20):4305–14.
Article108. Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997; 276:1404–7.
Article109. Zhang L, Wang H, Zhao Y, Wang J, Dubielecka PM, Zhuang S, et al. Myocyte-specific overexpressing HDAC4 promotes myocardial ischemia/reperfusion injury. Mol Med. 2018; 24:37.
Article110. Zou G, Zhong W, Wu F, Wang X, Liu L. Catalpol attenuates cardiomyocyte apoptosis in diabetic cardiomyopathy via Neat1/miR-140-5p/HDAC4 axis. Biochimie. 2019; 165:90–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Role of Histone Acetylation in Mesenchymal Stem Cell Differentiation
- HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases
- Epigenetic Modifications: Novel Therapeutic Approach for Thyroid Cancer
- SIRT1 in Type 2 Diabetes: Mechanisms and Therapeutic Potential
- Pathogenesis and Pathophysiology of Chronic Obstructive Pulmonary Disease (COPD)