Ann Lab Med.
2024 May;44(3):289-293. 10.3343/alm.2023.0256.
Neutralization Testing–based Immunogenicity Analysis of Recent Prevalent Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Sublineages
- Affiliations
-
- 1Division of Emerging Infectious Diseases, Bureau of Infectious Diseases Diagnosis Control, Korea Disease Control and Prevention Agency (KDCA), Cheongju, Korea
- 2College of Medicine and Medical Research Institute of Chungbuk National University, Cheongju, Korea
- KMID: 2555707
- DOI: http://doi.org/10.3343/alm.2023.0256
Abstract
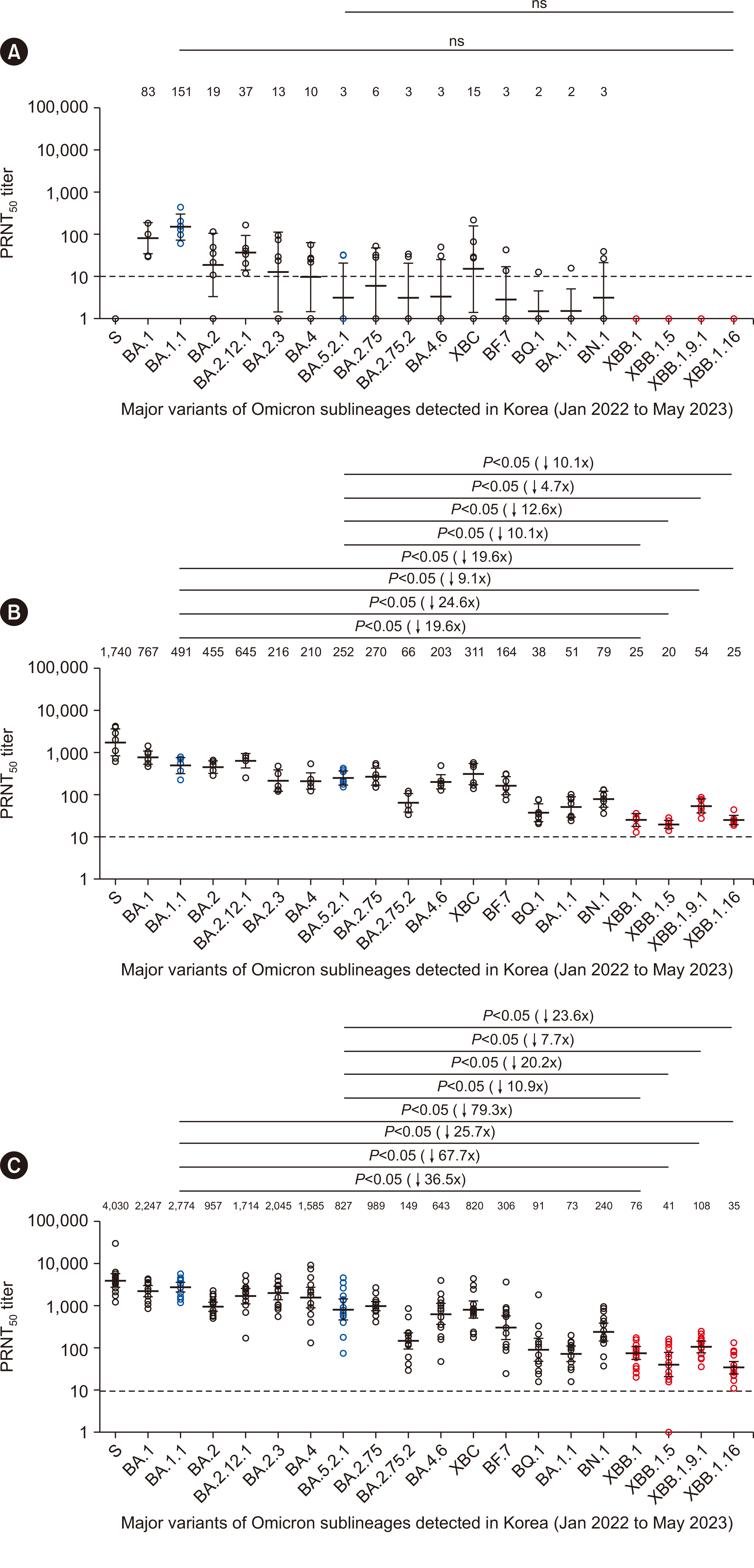
- Although WHO declared the end of the public health emergency for coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARSCoV-2), XBB lineages continue to evolve and emerge globally. In particular, XBB.1.5 and XBB.1.16 are raising concerns because of their high immune evasion, leading to apprehensions regarding vaccine efficacy reduction and potential reinfection. We aimed to investigate the COVID-19 outbreak in Korea and predict the likelihood of reinfection by testing neutralizing activity against live viruses from the S clade and 19 Omicron sublineages. We found a significant risk of infection with the currently prevalent XBB lineage for individuals who were either vaccinated early or infected during the initial Omicron outbreak. Vaccinated individuals were better equipped than unvaccinated individuals to produce neutralizing antibodies for other SARS-CoV-2 variants upon infection. Therefore, unvaccinated individuals do not easily develop neutralizing activity against other variants and face the highest risk of reinfection by the XBB lineage. Our study provides important information to facilitate the development of strategies for monitoring populations that would be the most susceptible to new COVID-19 outbreaks.
Figure
Reference
-
References
1. Statement on the fifteenth meeting of the IHR (2005) Emergency Committee on the COVID-19 pandemic. https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic. Updated on May 2023.2. KDCA press release. https://www.kdca.go.kr/board/board.es?mid=a20501010000&bid=0015&list_no=722664&cg_code=&act=view&nPage=2. Updated on May 2023.3. KDCA press release. https://www.kdca.go.kr/board/board.es?mid=a20501020000&bid=0015&list_no=723107&cg_code=C01&act=view&nPage=1. Updated on July 26, 2023.4. Qu P, Faraone JN, Evans JP, Zheng YM, Carlin C, Anghelina M, et al. 2023; Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants. Cell Rep. 42:112443. DOI: 10.1016/j.celrep.2023.112443. PMID: 37104089. PMCID: PMC10279473.5. Yamasoba D, Uriu K, Plianchaisuk A, Kosugi Y, Pan L, Zahradnik J, et al. 2023; Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect Dis. 23:655–6. DOI: 10.1016/S1473-3099(23)00278-5. PMID: 37148902.6. Kim HM, Lee EJ, O SW, Choi YJ, Lee H, Oh SJ, et al. 2023; A seroprevalence study on residents in a senior care facility with breakthrough SARS-CoV-2 Omicron infection. Viral Immunol. 36:203–8. DOI: 10.1089/vim.2022.0133. PMID: 36951666. PMCID: PMC10621659.7. Menegale F, Manica M, Zardini A, Guzzetta G, Marziano V, d'Andrea V, et al. 2023; Evaluation of waning of SARS-CoV-2 vaccine-induced immunity: a systematic review and meta-analysis. JAMA Netw Open. 6:e2310650. DOI: 10.1001/jamanetworkopen.2023.10650. PMID: 37133863. PMCID: PMC10157431.8. Harapan H, Ar Royan H, Tyas II, Nadira A, Abdi IF, Anwar S, et al. 2023; Waning anti-SARS-CoV-2 receptor-binding domain total antibody in CoronaVac-vaccinated individuals in Indonesia. F1000Res. 11:300. DOI: 10.12688/f1000research.109676.2. PMID: 37260419. PMCID: PMC10209622.9. Zhang L, Jiang L, Tian T, Li W, Pan Y, Wang Y. 2022; Efficacy and safety of COVID-19 vaccination in older adults: a systematic review and meta-analysis. Vaccines (Basel). 11:33. DOI: 10.3390/vaccines11010033. PMID: 36679878. PMCID: PMC9862835.10. Yang J, Hong W, Lei H, He C, Lei W, Zhou Y, et al. 2023; Low levels of neutralizing antibodies against XBB Omicron subvariants after BA.5 infection. Signal Transduct Target Ther. 8:252. DOI: 10.1038/s41392-023-01495-4. PMID: 37336889. PMCID: PMC10279763.11. Imai M, Ito M, Kiso M, Yamayoshi S, Uraki R, Fukushi S, et al. 2023; Efficacy of antiviral agents against Omicron subvariants BQ.1.1 and XBB. N Engl J Med. 388:89–91. DOI: 10.1056/NEJMc2214302. PMID: 36476720. PMCID: PMC9749618.12. Xia S, Jiao F, Wang L, Yu X, Lu T, Fu Y, et al. 2023; SARS-CoV-2 Omicron XBB subvariants exhibit enhanced fusogenicity and substantial immune evasion in elderly population, but high sensitivity to pan-coronavirus fusion inhibitors. J Med Virol. 95:e28641. DOI: 10.1002/jmv.28641. PMID: 36890632.13. Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, et al. 2023; Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 186:279–86.e8. DOI: 10.1016/j.cell.2022.12.018. PMID: 36580913. PMCID: PMC9747694.14. Wherry EJ, Barouch DH. 2022; T cell immunity to COVID-19 vaccines. Science. 377:821–2. DOI: 10.1126/science.add2897. PMID: 35981045.15. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al. 2022; mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 185:457–66.e4. DOI: 10.1016/j.cell.2021.12.033. PMID: 34995482. PMCID: PMC8733787.16. Dulipsingh L, Schaefer EJ, Wakefield D, Williams K, Halilovic A, Crowell R. 2023; Comparing SARS-CoV-2 neutralizing antibody levels in convalescent unvaccinated, convalescent vaccinated, and naive vaccinated subjects. Heliyon. 9:e17410. DOI: 10.1016/j.heliyon.2023.e17410. PMID: 37366522. PMCID: PMC10276490.17. Suarez Castillo MS, Khaoua H, Courtejoie N. 2022; Vaccine-induced and naturally-acquired protection against Omicron and Delta symptomatic infection and severe COVID-19 outcomes, France, December 2021 to January 2022. Euro Surveill. 27:2200250. DOI: 10.2807/1560-7917.ES.2022.27.16.2200250. PMID: 35451363. PMCID: PMC9027152.18. Bobrovitz N, Ware H, Ma X, Li Z, Hosseini R, Cao C, et al. 2023; Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect Dis. 23:556–67. DOI: 10.1016/S1473-3099(22)00801-5. PMID: 36681084.19. Guide to quarantine measures following the lowering of the COVID-19 alert level [in Korean]. https://www.kdca.go.kr/gallery.es?mid=a20503020000&bid=0003&b_list=9&act=view&list_no=146120&nPage=1&vlist_no_npage=1&keyField=&keyWord=&orderby=. Updated on May 2023.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Replication kinetics and infectivity of SARS-CoV-2 Omicron variant sublineages recovered in the Republic of Korea
- Increased viral load in patients infected with severe acute respiratory syndrome coronavirus 2 Omicron variant in the Republic of Korea
- Humoral Immune Response of Heterologous ChAdOx1 nCoV-19 and mRNA-1273 Prime-Boost Vaccination against SARS-CoV-2 Variants in Korea
- Neutralizing Activity and T-Cell Responses Against Wild Type SARSCoV-2 Virus and Omicron BA.5 Variant After Ancestral SARS-CoV-2 Vaccine Booster Dose in PLWH Receiving ART Based on CD4 T-Cell Count
- Recombinant proteins of spike protein of SARS-CoV-2 with the Omicron receptor-binding domain induce production of highly Omicron-specific neutralizing antibodies